You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003201_00521
You are here: Home > Sequence: MGYG000003201_00521
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
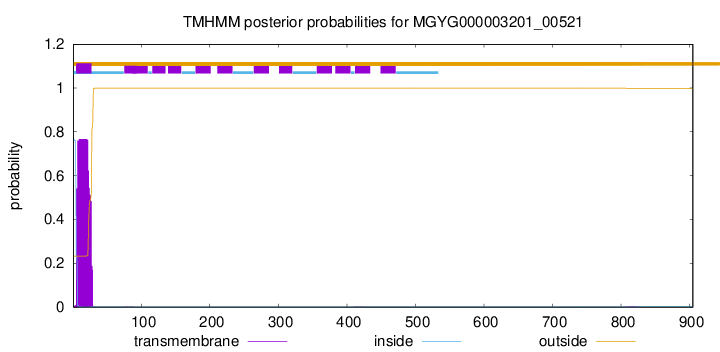
TMHMM annotations
Basic Information help
| Species | CAG-873 sp900759825 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; CAG-873; CAG-873 sp900759825 | |||||||||||
| CAZyme ID | MGYG000003201_00521 | |||||||||||
| CAZy Family | PL15 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1737; End: 4454 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL15 | 550 | 710 | 4.7e-41 | 0.9776119402985075 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam16332 | DUF4962 | 8.79e-53 | 61 | 520 | 8 | 473 | Domain of unknown function (DUF4962). This family consists of uncharacterized proteins around 870 residues in length and is mainly found in various Bacteroides species. The function of this protein is unknown. |
| pfam07940 | Hepar_II_III | 6.16e-11 | 533 | 763 | 4 | 203 | Heparinase II/III-like protein. This family features sequences that are similar to a region of the Flavobacterium heparinum proteins heparinase II and heparinase III. The former is known to degrade heparin and heparin sulphate, whereas the latter predominantly degrades heparin sulphate. Both are secreted into the periplasmic space upon induction with heparin. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ARS40195.1 | 1.02e-294 | 2 | 899 | 11 | 916 |
| AKP51277.1 | 1.74e-87 | 64 | 856 | 67 | 820 |
| BCI64400.1 | 8.76e-87 | 61 | 869 | 39 | 841 |
| QUE51929.1 | 2.16e-86 | 61 | 897 | 36 | 819 |
| QIU94214.1 | 1.71e-85 | 61 | 898 | 49 | 843 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6LJA_A | 7.57e-74 | 39 | 891 | 18 | 870 | ChainA, Heparinase II/III-like protein [Bacteroides intestinalis DSM 17393] |
| 6LJL_A | 8.40e-72 | 39 | 891 | 18 | 870 | ChainA, Heparinase II/III-like protein [Bacteroides intestinalis DSM 17393] |
| 3A0O_A | 6.55e-17 | 66 | 653 | 28 | 619 | Crystalstructure of alginate lyase from Agrobacterium tumefaciens C58 [Agrobacterium fabrum str. C58],3A0O_B Crystal structure of alginate lyase from Agrobacterium tumefaciens C58 [Agrobacterium fabrum str. C58] |
| 3AFL_A | 1.02e-15 | 66 | 653 | 28 | 619 | Crystalstructure of exotype alginate lyase Atu3025 H531A complexed with alginate trisaccharide [Agrobacterium fabrum str. C58] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000219 | 0.999143 | 0.000165 | 0.000151 | 0.000147 | 0.000133 |