You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003205_00075
You are here: Home > Sequence: MGYG000003205_00075
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-312 sp900760055 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Verrucomicrobiae; Opitutales; CAG-312; CAG-312; CAG-312 sp900760055 | |||||||||||
| CAZyme ID | MGYG000003205_00075 | |||||||||||
| CAZy Family | GH39 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 48038; End: 49963 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH39 | 73 | 295 | 1.5e-27 | 0.5846867749419954 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3693 | XynA | 1.56e-08 | 63 | 247 | 65 | 258 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| pfam02449 | Glyco_hydro_42 | 1.83e-08 | 40 | 136 | 6 | 101 | Beta-galactosidase. This group of beta-galactosidase enzymes belong to the glycosyl hydrolase 42 family. The enzyme catalyzes the hydrolysis of terminal, non-reducing terminal beta-D-galactosidase residues. |
| COG1874 | GanA | 2.01e-07 | 32 | 129 | 18 | 122 | Beta-galactosidase GanA [Carbohydrate transport and metabolism]. |
| smart00633 | Glyco_10 | 2.07e-05 | 67 | 247 | 3 | 194 | Glycosyl hydrolase family 10. |
| pfam00232 | Glyco_hydro_1 | 0.002 | 35 | 213 | 54 | 255 | Glycosyl hydrolase family 1. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVM44885.1 | 2.69e-119 | 9 | 637 | 6 | 614 |
| QDU57372.1 | 4.89e-84 | 30 | 634 | 28 | 625 |
| AVM44901.1 | 5.04e-57 | 30 | 615 | 25 | 608 |
| AHF90941.1 | 2.05e-42 | 30 | 605 | 232 | 830 |
| ACZ43011.1 | 5.91e-34 | 22 | 251 | 294 | 530 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5JVK_A | 2.50e-17 | 33 | 251 | 107 | 345 | Structuralinsights into a family 39 glycoside hydrolase from the gut symbiont Bacteroides cellulosilyticus WH2. [Bacteroides cellulosilyticus],5JVK_B Structural insights into a family 39 glycoside hydrolase from the gut symbiont Bacteroides cellulosilyticus WH2. [Bacteroides cellulosilyticus],5JVK_C Structural insights into a family 39 glycoside hydrolase from the gut symbiont Bacteroides cellulosilyticus WH2. [Bacteroides cellulosilyticus] |
| 3WUB_A | 2.41e-06 | 65 | 225 | 47 | 211 | Thewild type crystal structure of b-1,4-Xylanase (XynAS9) from Streptomyces sp. 9 [Streptomyces sp.],3WUE_A The wild type crystal structure of b-1,4-Xylanase (XynAS9) with xylobiose from Streptomyces sp. 9 [Streptomyces sp.] |
| 3WUF_A | 2.41e-06 | 65 | 225 | 47 | 211 | Themutant crystal structure of b-1,4-Xylanase (XynAS9_V43P/G44E) from Streptomyces sp. 9 [Streptomyces sp.],3WUG_A The mutant crystal structure of b-1,4-Xylanase (XynAS9_V43P/G44E) with xylobiose from Streptomyces sp. 9 [Streptomyces sp.] |
| 6Z1H_A | 2.74e-06 | 27 | 180 | 52 | 213 | ChainA, ANCESTRAL RECONSTRUCTED GLYCOSIDASE [synthetic construct],6Z1H_B Chain B, ANCESTRAL RECONSTRUCTED GLYCOSIDASE [synthetic construct],6Z1M_A Chain A, Ancestral reconstructed glycosidase [synthetic construct],6Z1M_B Chain B, Ancestral reconstructed glycosidase [synthetic construct],6Z1M_C Chain C, Ancestral reconstructed glycosidase [synthetic construct] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as LIPO
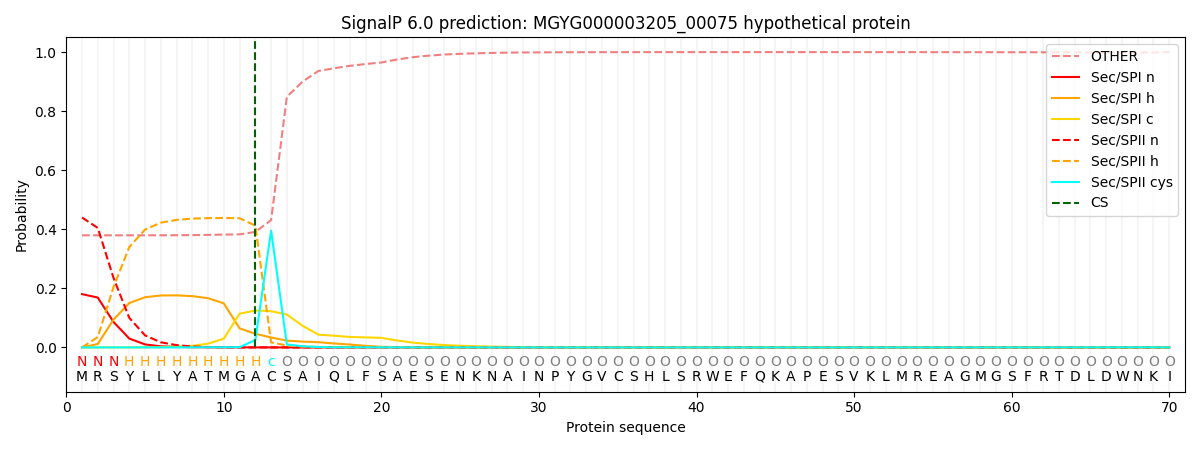
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.387348 | 0.170035 | 0.441954 | 0.000214 | 0.000169 | 0.000272 |
