You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003205_00076
You are here: Home > Sequence: MGYG000003205_00076
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
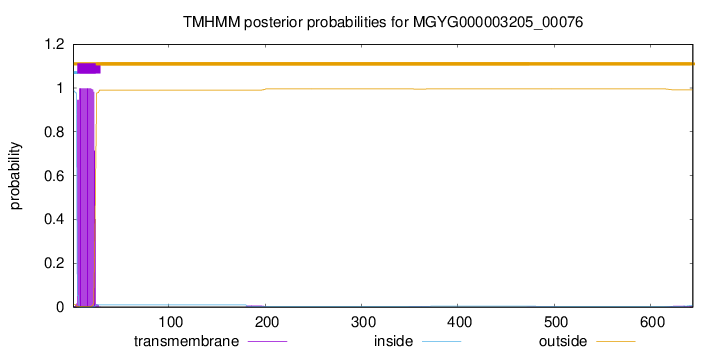
TMHMM annotations
Basic Information help
| Species | CAG-312 sp900760055 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Verrucomicrobiae; Opitutales; CAG-312; CAG-312; CAG-312 sp900760055 | |||||||||||
| CAZyme ID | MGYG000003205_00076 | |||||||||||
| CAZy Family | GH39 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 50033; End: 51967 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH39 | 75 | 262 | 1.2e-25 | 0.5104408352668214 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02449 | Glyco_hydro_42 | 2.91e-05 | 55 | 110 | 16 | 72 | Beta-galactosidase. This group of beta-galactosidase enzymes belong to the glycosyl hydrolase 42 family. The enzyme catalyzes the hydrolysis of terminal, non-reducing terminal beta-D-galactosidase residues. |
| COG1874 | GanA | 4.82e-05 | 37 | 110 | 18 | 94 | Beta-galactosidase GanA [Carbohydrate transport and metabolism]. |
| COG3693 | XynA | 1.02e-04 | 72 | 270 | 69 | 276 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| COG2723 | BglB | 3.89e-04 | 55 | 175 | 65 | 197 | Beta-glucosidase/6-phospho-beta-glucosidase/beta-galactosidase [Carbohydrate transport and metabolism]. |
| pfam11790 | Glyco_hydro_cc | 4.02e-04 | 199 | 263 | 124 | 179 | Glycosyl hydrolase catalytic core. This family is probably a glycosyl hydrolase, and is conserved in fungi and some Proteobacteria. The pombe member is annotated as being from IPR013781. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVM44885.1 | 2.58e-113 | 35 | 614 | 27 | 590 |
| QDU57372.1 | 9.89e-75 | 9 | 638 | 7 | 624 |
| AVM44901.1 | 1.15e-48 | 23 | 591 | 13 | 585 |
| AHF90941.1 | 1.44e-39 | 23 | 591 | 216 | 804 |
| ACZ43011.1 | 1.07e-32 | 29 | 296 | 296 | 570 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5JVK_A | 2.88e-11 | 31 | 256 | 85 | 345 | Structuralinsights into a family 39 glycoside hydrolase from the gut symbiont Bacteroides cellulosilyticus WH2. [Bacteroides cellulosilyticus],5JVK_B Structural insights into a family 39 glycoside hydrolase from the gut symbiont Bacteroides cellulosilyticus WH2. [Bacteroides cellulosilyticus],5JVK_C Structural insights into a family 39 glycoside hydrolase from the gut symbiont Bacteroides cellulosilyticus WH2. [Bacteroides cellulosilyticus] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER
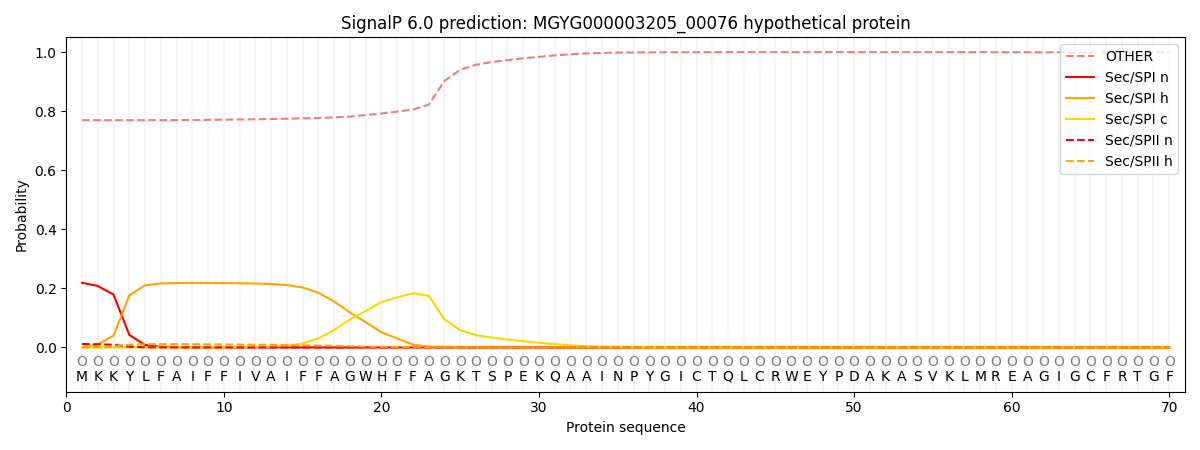
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.779673 | 0.207326 | 0.012227 | 0.000250 | 0.000164 | 0.000364 |