You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003205_00500
You are here: Home > Sequence: MGYG000003205_00500
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-312 sp900760055 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Verrucomicrobiae; Opitutales; CAG-312; CAG-312; CAG-312 sp900760055 | |||||||||||
| CAZyme ID | MGYG000003205_00500 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | Anti-sigma-I factor RsgI6 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 88343; End: 89800 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 100 | 424 | 2.2e-37 | 0.9570957095709571 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| smart00633 | Glyco_10 | 4.52e-25 | 141 | 422 | 3 | 260 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 4.64e-25 | 141 | 425 | 69 | 337 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QMW04636.1 | 6.46e-125 | 41 | 475 | 14 | 448 |
| QNL52536.1 | 7.71e-112 | 50 | 465 | 16 | 418 |
| AEI51978.1 | 4.87e-109 | 54 | 478 | 24 | 435 |
| QIP12711.1 | 1.20e-107 | 49 | 474 | 19 | 435 |
| QHV94149.1 | 1.35e-106 | 49 | 474 | 19 | 435 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5AY7_A | 4.80e-09 | 172 | 431 | 78 | 349 | Apsychrophilic glycoside hydrolase family 10 endo-beta-1,4-xylanase [Aegilops speltoides subsp. speltoides],5AY7_B A psychrophilic glycoside hydrolase family 10 endo-beta-1,4-xylanase [Aegilops speltoides subsp. speltoides],5D4Y_A A psychrophilic glycoside hydrolase family 10 endo-beta-1,4-xylanase [environmental samples],5D4Y_B A psychrophilic glycoside hydrolase family 10 endo-beta-1,4-xylanase [environmental samples] |
| 3EMC_A | 1.00e-07 | 163 | 356 | 55 | 241 | Crystalstructure of XynB, an intracellular xylanase from Paenibacillus barcinonensis [Paenibacillus barcinonensis],3EMQ_A Crystal structure of xilanase XynB from Paenibacillus barcelonensis complexed with an inhibitor [Paenibacillus barcinonensis],3EMZ_A Crystal structure of xylanase XynB from Paenibacillus barcinonensis complexed with a conduramine derivative [Paenibacillus barcinonensis] |
| 6FHE_A | 1.84e-07 | 116 | 398 | 34 | 305 | Highlyactive enzymes by automated modular backbone assembly and sequence design [synthetic construct] |
| 1XYZ_A | 4.46e-07 | 172 | 429 | 90 | 344 | ChainA, 1,4-BETA-D-XYLAN-XYLANOHYDROLASE [Acetivibrio thermocellus],1XYZ_B Chain B, 1,4-BETA-D-XYLAN-XYLANOHYDROLASE [Acetivibrio thermocellus] |
| 6D5C_A | 5.99e-07 | 166 | 422 | 80 | 344 | Structureof Caldicellulosiruptor danielii GH10 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii],6D5C_B Structure of Caldicellulosiruptor danielii GH10 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii],6D5C_C Structure of Caldicellulosiruptor danielii GH10 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A3DH97 | 3.79e-23 | 69 | 475 | 388 | 747 | Anti-sigma-I factor RsgI6 OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=rsgI6 PE=1 SV=1 |
| Q12603 | 1.10e-08 | 77 | 355 | 18 | 262 | Beta-1,4-xylanase OS=Dictyoglomus thermophilum OX=14 GN=xynA PE=3 SV=1 |
| Q60041 | 1.05e-07 | 163 | 432 | 79 | 342 | Endo-1,4-beta-xylanase B OS=Thermotoga neapolitana OX=2337 GN=xynB PE=3 SV=1 |
| P23557 | 2.10e-07 | 175 | 358 | 31 | 219 | Putative endo-1,4-beta-xylanase OS=Caldicellulosiruptor saccharolyticus OX=44001 PE=3 SV=1 |
| D5EY13 | 4.45e-07 | 100 | 425 | 38 | 366 | Endo-1,4-beta-xylanase/feruloyl esterase OS=Prevotella ruminicola (strain ATCC 19189 / JCM 8958 / 23) OX=264731 GN=xyn10D-fae1A PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
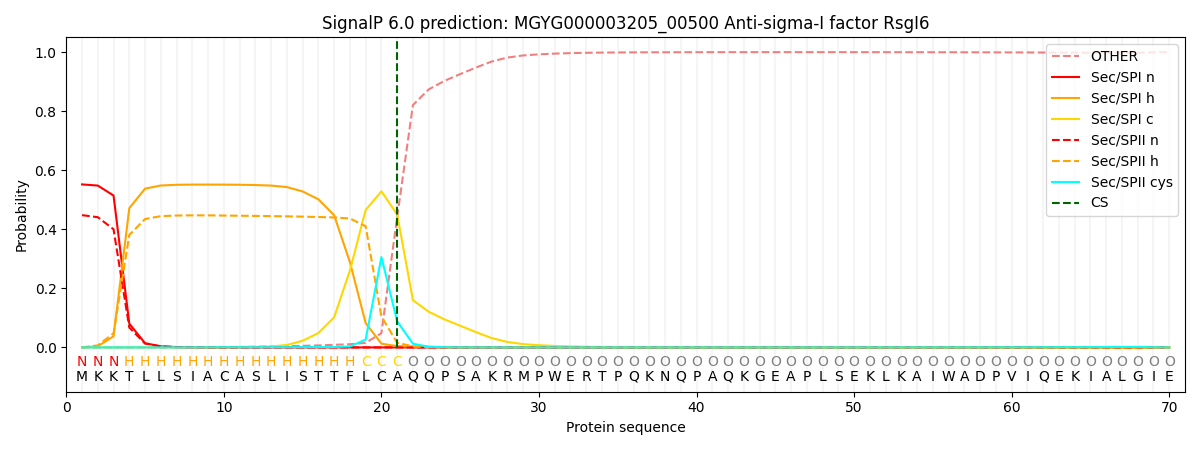
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000652 | 0.542868 | 0.455705 | 0.000323 | 0.000226 | 0.000196 |
