You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003205_01340
You are here: Home > Sequence: MGYG000003205_01340
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-312 sp900760055 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Verrucomicrobiae; Opitutales; CAG-312; CAG-312; CAG-312 sp900760055 | |||||||||||
| CAZyme ID | MGYG000003205_01340 | |||||||||||
| CAZy Family | GH106 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 24315; End: 27524 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH106 | 42 | 835 | 9.9e-64 | 0.9368932038834952 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam17132 | Glyco_hydro_106 | 4.96e-13 | 190 | 563 | 401 | 755 | alpha-L-rhamnosidase. |
| cd01210 | PTB_EPS8 | 0.003 | 549 | 592 | 28 | 78 | Epidermal growth factor receptor kinase substrate (EPS8)-like Phosphotyrosine-binding (PTB) domain. EPS8 is a regulator of Rac signaling. It consists of a PTB and an SH3 domain. PTB domains have a common PH-like fold and are found in various eukaryotic signaling molecules. This domain was initially shown to binds peptides with a NPXY motif with differing requirements for phosphorylation of the tyrosine, although more recent studies have found that some types of PTB domains can bind to peptides lack tyrosine residues altogether. In contrast to SH2 domains, which recognize phosphotyrosine and adjacent carboxy-terminal residues, PTB-domain binding specificity is conferred by residues amino-terminal to the phosphotyrosine. PTB domains are classified into three groups: phosphotyrosine-dependent Shc-like, phosphotyrosine-dependent IRS-like, and phosphotyrosine-independent Dab-like PTB domains. This cd is part of the Dab-like subgroup. |
| smart00798 | AICARFT_IMPCHas | 0.005 | 221 | 261 | 126 | 165 | AICARFT/IMPCHase bienzyme. This is a family of bifunctional enzymes catalysing the last two steps in de novo purine biosynthesis. The bifunctional enzyme is found in both prokaryotes and eukaryotes. The second last step is catalysed by 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase (AICARFT), this enzyme catalyses the formylation of AICAR with 10-formyl-tetrahydrofolate to yield FAICAR and tetrahydrofolate. The last step is catalysed by IMP (Inosine monophosphate) cyclohydrolase (IMPCHase), cyclizing FAICAR (5-formylaminoimidazole-4-carboxamide ribonucleotide) to IMP. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| SCM56672.1 | 1.05e-161 | 41 | 1063 | 29 | 1024 |
| QMV43982.1 | 2.85e-160 | 44 | 1061 | 11 | 1022 |
| QHQ61014.1 | 4.85e-151 | 44 | 1058 | 5 | 1014 |
| AEE96868.1 | 1.00e-142 | 42 | 1058 | 5 | 1007 |
| BCG53829.1 | 1.90e-141 | 37 | 1052 | 37 | 1055 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| T2KNA8 | 1.08e-11 | 420 | 1026 | 486 | 1104 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22040 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
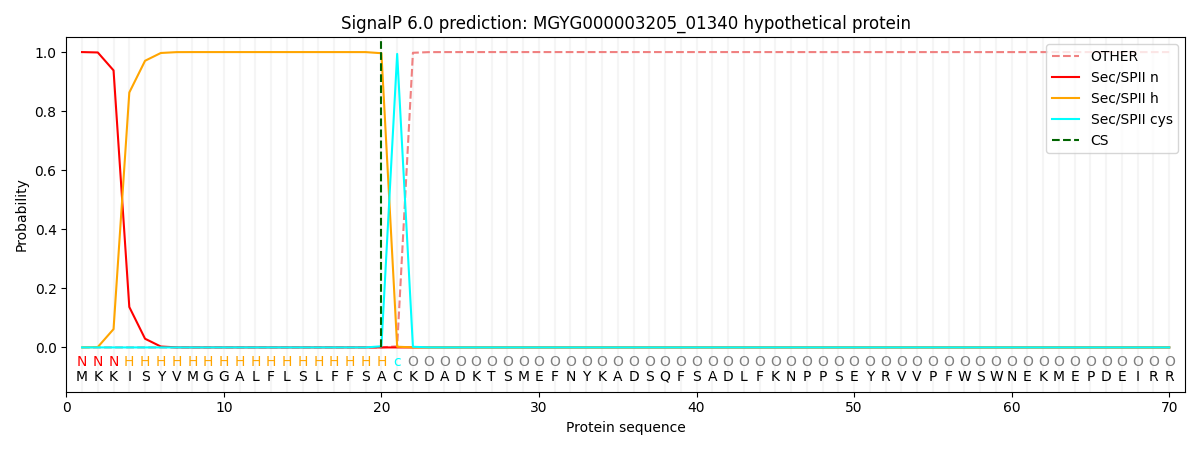
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000044 | 0.000000 | 0.000000 | 0.000000 |
