You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003205_01975
You are here: Home > Sequence: MGYG000003205_01975
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-312 sp900760055 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Verrucomicrobiae; Opitutales; CAG-312; CAG-312; CAG-312 sp900760055 | |||||||||||
| CAZyme ID | MGYG000003205_01975 | |||||||||||
| CAZy Family | GH42 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 55778; End: 58480 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH42 | 297 | 484 | 9.9e-19 | 0.4555256064690027 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02449 | Glyco_hydro_42 | 1.19e-15 | 298 | 470 | 72 | 229 | Beta-galactosidase. This group of beta-galactosidase enzymes belong to the glycosyl hydrolase 42 family. The enzyme catalyzes the hydrolysis of terminal, non-reducing terminal beta-D-galactosidase residues. |
| COG1874 | GanA | 1.56e-06 | 298 | 525 | 94 | 293 | Beta-galactosidase GanA [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ASV74990.1 | 8.11e-128 | 30 | 895 | 33 | 872 |
| QHI69172.1 | 2.63e-50 | 273 | 872 | 307 | 918 |
| SDT33465.1 | 7.06e-48 | 274 | 869 | 132 | 736 |
| AVM44099.1 | 2.99e-23 | 271 | 625 | 236 | 588 |
| QIK54901.1 | 1.85e-11 | 298 | 544 | 109 | 360 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6Y2K_A | 7.42e-09 | 298 | 447 | 77 | 223 | ChainA, beta-galactosidase [Marinomonas sp. ef1] |
| 4UCF_A | 1.31e-08 | 250 | 478 | 45 | 263 | Crystalstructure of Bifidobacterium bifidum beta-galactosidase in complex with alpha-galactose [Bifidobacterium bifidum S17],4UCF_B Crystal structure of Bifidobacterium bifidum beta-galactosidase in complex with alpha-galactose [Bifidobacterium bifidum S17],4UCF_C Crystal structure of Bifidobacterium bifidum beta-galactosidase in complex with alpha-galactose [Bifidobacterium bifidum S17],4UZS_A Crystal structure of Bifidobacterium bifidum beta-galactosidase [Bifidobacterium bifidum S17],4UZS_B Crystal structure of Bifidobacterium bifidum beta-galactosidase [Bifidobacterium bifidum S17],4UZS_C Crystal structure of Bifidobacterium bifidum beta-galactosidase [Bifidobacterium bifidum S17] |
| 1KWG_A | 8.63e-08 | 298 | 481 | 76 | 245 | Crystalstructure of Thermus thermophilus A4 beta-galactosidase [Thermus thermophilus],1KWK_A Crystal structure of Thermus thermophilus A4 beta-galactosidase in complex with galactose [Thermus thermophilus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A1A399 | 5.51e-08 | 250 | 478 | 45 | 263 | Beta-galactosidase BgaB OS=Bifidobacterium adolescentis (strain ATCC 15703 / DSM 20083 / NCTC 11814 / E194a) OX=367928 GN=bgaB PE=1 SV=2 |
| Q8GEA9 | 6.93e-08 | 298 | 481 | 76 | 245 | Beta-galactosidase BgaA OS=Thermus sp. OX=275 GN=bgaA PE=1 SV=1 |
| D4QFE7 | 7.19e-08 | 250 | 478 | 45 | 263 | Beta-galactosidase BbgII OS=Bifidobacterium bifidum OX=1681 GN=bbgII PE=3 SV=1 |
| O69315 | 4.73e-07 | 298 | 481 | 76 | 245 | Beta-galactosidase OS=Thermus thermophilus OX=274 PE=1 SV=1 |
| Q93GI5 | 8.47e-07 | 250 | 471 | 44 | 255 | Beta-galactosidase III OS=Bifidobacterium longum subsp. infantis OX=1682 GN=beta-galIII PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
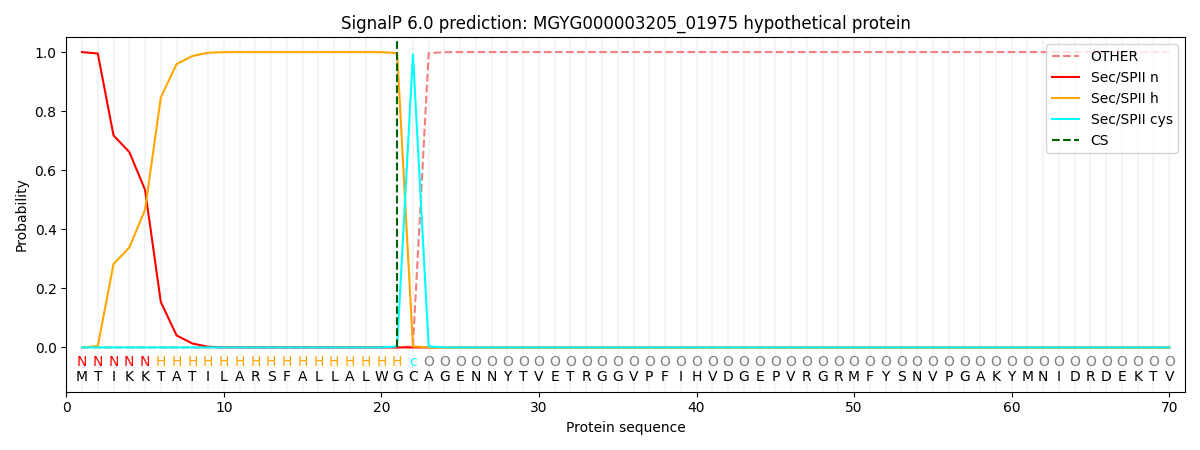
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000001 | 1.000051 | 0.000000 | 0.000000 | 0.000000 |
