You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003205_02025
You are here: Home > Sequence: MGYG000003205_02025
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-312 sp900760055 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Verrucomicrobiae; Opitutales; CAG-312; CAG-312; CAG-312 sp900760055 | |||||||||||
| CAZyme ID | MGYG000003205_02025 | |||||||||||
| CAZy Family | GH42 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 9389; End: 11965 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH42 | 196 | 435 | 1.8e-18 | 0.6091644204851752 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02449 | Glyco_hydro_42 | 5.70e-16 | 196 | 441 | 9 | 247 | Beta-galactosidase. This group of beta-galactosidase enzymes belong to the glycosyl hydrolase 42 family. The enzyme catalyzes the hydrolysis of terminal, non-reducing terminal beta-D-galactosidase residues. |
| COG1874 | GanA | 5.70e-10 | 199 | 433 | 32 | 254 | Beta-galactosidase GanA [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ASV74990.1 | 9.41e-130 | 24 | 854 | 32 | 873 |
| QHI69172.1 | 5.67e-52 | 224 | 834 | 298 | 922 |
| SDT33465.1 | 1.20e-51 | 209 | 827 | 89 | 736 |
| AVM44099.1 | 2.41e-25 | 226 | 658 | 231 | 669 |
| AYC38161.1 | 3.66e-08 | 196 | 429 | 25 | 242 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4UNI_A | 2.52e-07 | 200 | 422 | 40 | 250 | beta-(1,6)-galactosidasefrom Bifidobacterium animalis subsp. lactis Bl-04 in complex with galactose [Bifidobacterium animalis subsp. lactis Bl-04],4UNI_B beta-(1,6)-galactosidase from Bifidobacterium animalis subsp. lactis Bl-04 in complex with galactose [Bifidobacterium animalis subsp. lactis Bl-04],4UNI_C beta-(1,6)-galactosidase from Bifidobacterium animalis subsp. lactis Bl-04 in complex with galactose [Bifidobacterium animalis subsp. lactis Bl-04] |
| 4UOQ_A | 2.52e-07 | 200 | 422 | 40 | 250 | Nucleophilemutant (E324A) of beta-(1,6)-galactosidase from Bifidobacterium animalis subsp. lactis Bl-04 [Bifidobacterium animalis subsp. lactis Bl-04],4UOQ_B Nucleophile mutant (E324A) of beta-(1,6)-galactosidase from Bifidobacterium animalis subsp. lactis Bl-04 [Bifidobacterium animalis subsp. lactis Bl-04],4UOQ_C Nucleophile mutant (E324A) of beta-(1,6)-galactosidase from Bifidobacterium animalis subsp. lactis Bl-04 [Bifidobacterium animalis subsp. lactis Bl-04],4UOZ_A beta-(1,6)-galactosidase from Bifidobacterium animalis subsp. lactis Bl-04 nucleophile mutant E324A in complex with galactose [Bifidobacterium animalis subsp. lactis Bl-04],4UOZ_B beta-(1,6)-galactosidase from Bifidobacterium animalis subsp. lactis Bl-04 nucleophile mutant E324A in complex with galactose [Bifidobacterium animalis subsp. lactis Bl-04],4UOZ_C beta-(1,6)-galactosidase from Bifidobacterium animalis subsp. lactis Bl-04 nucleophile mutant E324A in complex with galactose [Bifidobacterium animalis subsp. lactis Bl-04] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
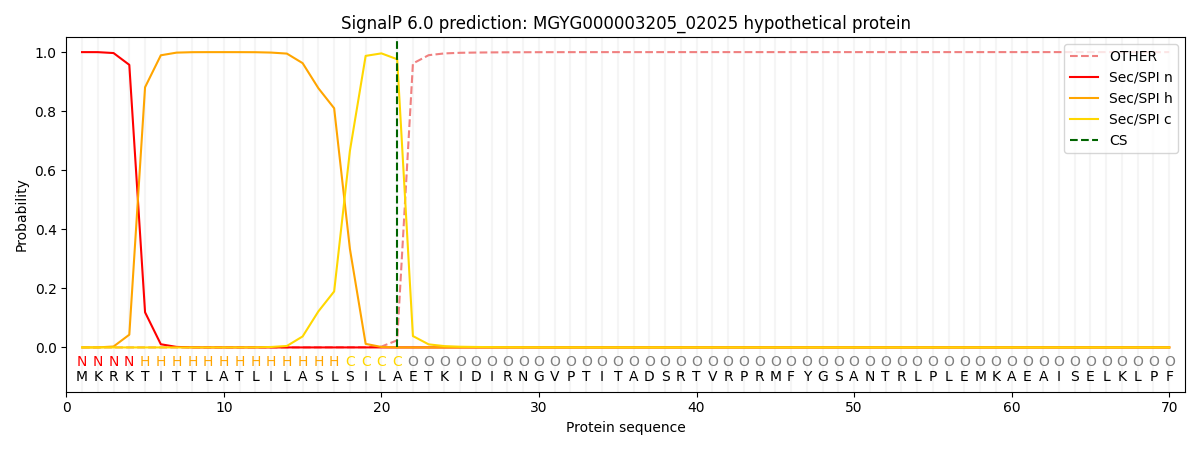
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000510 | 0.998474 | 0.000405 | 0.000205 | 0.000205 | 0.000184 |
