You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003215_01342
You are here: Home > Sequence: MGYG000003215_01342
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
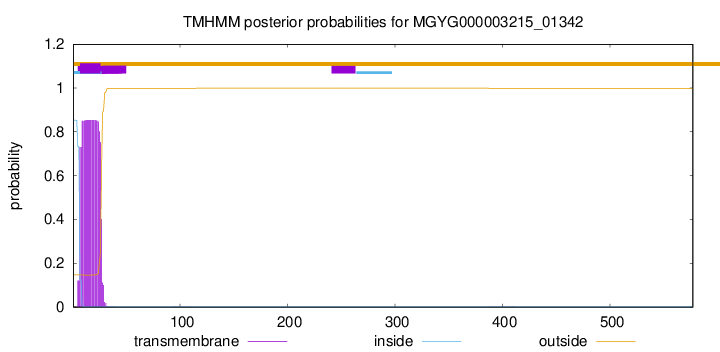
TMHMM annotations
Basic Information help
| Species | 1XD42-69 sp014287635 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; 1XD42-69; 1XD42-69 sp014287635 | |||||||||||
| CAZyme ID | MGYG000003215_01342 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3049; End: 4782 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH73 | 448 | 575 | 6.4e-21 | 0.9765625 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3401 | FN3 | 4.62e-14 | 83 | 289 | 126 | 328 | Fibronectin type 3 domain [General function prediction only]. |
| pfam01832 | Glucosaminidase | 1.26e-07 | 448 | 524 | 1 | 81 | Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase. This family includes Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase EC:3.2.1.96. As well as the flageller protein J that has been shown to hydrolyze peptidoglycan. |
| cd00063 | FN3 | 3.18e-05 | 128 | 214 | 8 | 93 | Fibronectin type 3 domain; One of three types of internal repeats found in the plasma protein fibronectin. Its tenth fibronectin type III repeat contains an RGD cell recognition sequence in a flexible loop between 2 strands. Approximately 2% of all animal proteins contain the FN3 repeat; including extracellular and intracellular proteins, membrane spanning cytokine receptors, growth hormone receptors, tyrosine phosphatase receptors, and adhesion molecules. FN3-like domains are also found in bacterial glycosyl hydrolases. |
| COG3401 | FN3 | 4.33e-05 | 246 | 390 | 102 | 242 | Fibronectin type 3 domain [General function prediction only]. |
| COG3401 | FN3 | 2.95e-04 | 42 | 190 | 178 | 322 | Fibronectin type 3 domain [General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| VCV20908.1 | 3.08e-108 | 42 | 574 | 931 | 1463 |
| CBL13779.1 | 1.91e-106 | 42 | 574 | 926 | 1458 |
| CBL07763.1 | 3.39e-105 | 42 | 574 | 925 | 1457 |
| QOY61400.1 | 7.85e-56 | 409 | 571 | 16 | 181 |
| QCU04040.1 | 2.12e-43 | 407 | 575 | 55 | 227 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3MPC_A | 5.39e-06 | 218 | 301 | 9 | 95 | ChainA, Fn3-like protein [Acetivibrio thermocellus ATCC 27405],3MPC_B Chain B, Fn3-like protein [Acetivibrio thermocellus ATCC 27405] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O51481 | 5.09e-23 | 449 | 577 | 73 | 198 | Uncharacterized protein BB_0531 OS=Borreliella burgdorferi (strain ATCC 35210 / DSM 4680 / CIP 102532 / B31) OX=224326 GN=BB_0531 PE=3 SV=1 |
| P38939 | 5.23e-09 | 42 | 287 | 940 | 1234 | Amylopullulanase OS=Thermoanaerobacter pseudethanolicus (strain ATCC 33223 / 39E) OX=340099 GN=apu PE=1 SV=2 |
| P16950 | 3.57e-08 | 42 | 321 | 941 | 1265 | Amylopullulanase OS=Thermoanaerobacter thermohydrosulfuricus OX=1516 GN=apu PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
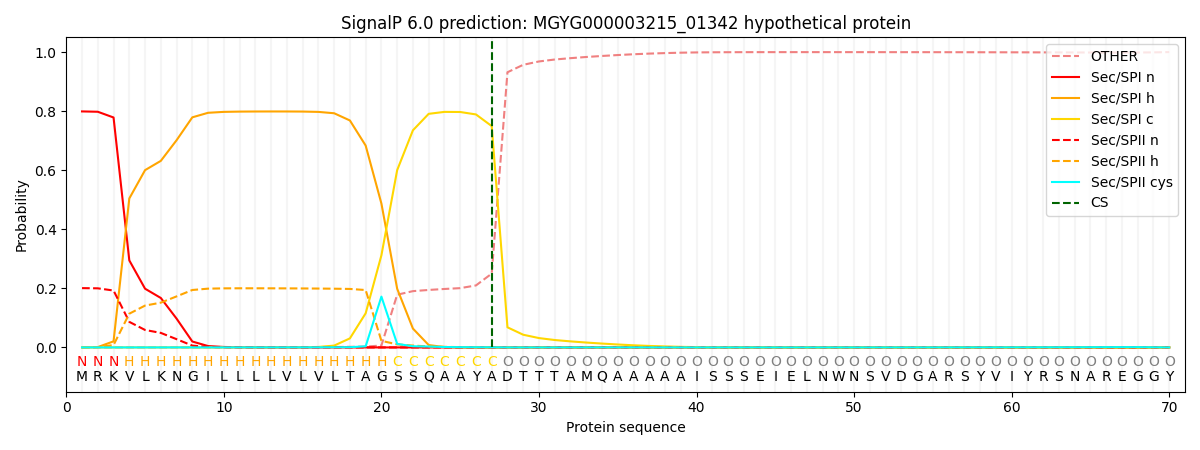
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000520 | 0.790935 | 0.207509 | 0.000489 | 0.000289 | 0.000229 |