You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003224_01329
You are here: Home > Sequence: MGYG000003224_01329
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | SFFH01 sp900542445 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia_A; Christensenellales; CAG-74; SFFH01; SFFH01 sp900542445 | |||||||||||
| CAZyme ID | MGYG000003224_01329 | |||||||||||
| CAZy Family | GT39 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 63049; End: 66969 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT39 | 871 | 1097 | 1.6e-48 | 0.9237668161434978 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam16192 | PMT_4TMC | 4.65e-28 | 1112 | 1301 | 1 | 198 | C-terminal four TMM region of protein-O-mannosyltransferase. PMT_4TMC is the C-terminal four membrane-pass region of protein-O-mannosyltransferases and similar enzymes. |
| COG1928 | PMT1 | 4.11e-21 | 1118 | 1303 | 505 | 693 | Dolichyl-phosphate-mannose--protein O-mannosyl transferase [Posttranslational modification, protein turnover, chaperones]. |
| COG5542 | COG5542 | 2.71e-06 | 302 | 598 | 92 | 377 | Mannosyltransferase related to Gpi18 [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QTE68569.1 | 7.24e-240 | 10 | 1294 | 11 | 1391 |
| CQR52353.1 | 1.22e-222 | 36 | 1304 | 29 | 1274 |
| AIQ56104.1 | 2.06e-221 | 36 | 1304 | 29 | 1278 |
| AIQ39188.1 | 8.02e-221 | 36 | 1304 | 29 | 1278 |
| QSF45552.1 | 1.34e-220 | 12 | 1304 | 9 | 1272 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| L8F4Z2 | 1.00e-21 | 869 | 1303 | 59 | 515 | Probable dolichyl-phosphate-mannose--protein mannosyltransferase OS=Mycolicibacterium smegmatis (strain MKD8) OX=1214915 GN=pmt PE=3 SV=1 |
| P9WN04 | 2.21e-18 | 900 | 1303 | 102 | 521 | Probable dolichyl-phosphate-mannose--protein mannosyltransferase OS=Mycobacterium tuberculosis (strain CDC 1551 / Oshkosh) OX=83331 GN=pmt PE=3 SV=2 |
| P9WN05 | 2.21e-18 | 900 | 1303 | 102 | 521 | Probable dolichyl-phosphate-mannose--protein mannosyltransferase OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=pmt PE=1 SV=2 |
| Q8NRZ6 | 9.95e-15 | 900 | 1303 | 99 | 519 | Probable dolichyl-phosphate-mannose--protein mannosyltransferase OS=Corynebacterium glutamicum (strain ATCC 13032 / DSM 20300 / BCRC 11384 / JCM 1318 / LMG 3730 / NCIMB 10025) OX=196627 GN=pmt PE=3 SV=1 |
| Q5ACU3 | 6.67e-10 | 1108 | 1303 | 506 | 716 | Dolichyl-phosphate-mannose--protein mannosyltransferase 5 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=PMT5 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
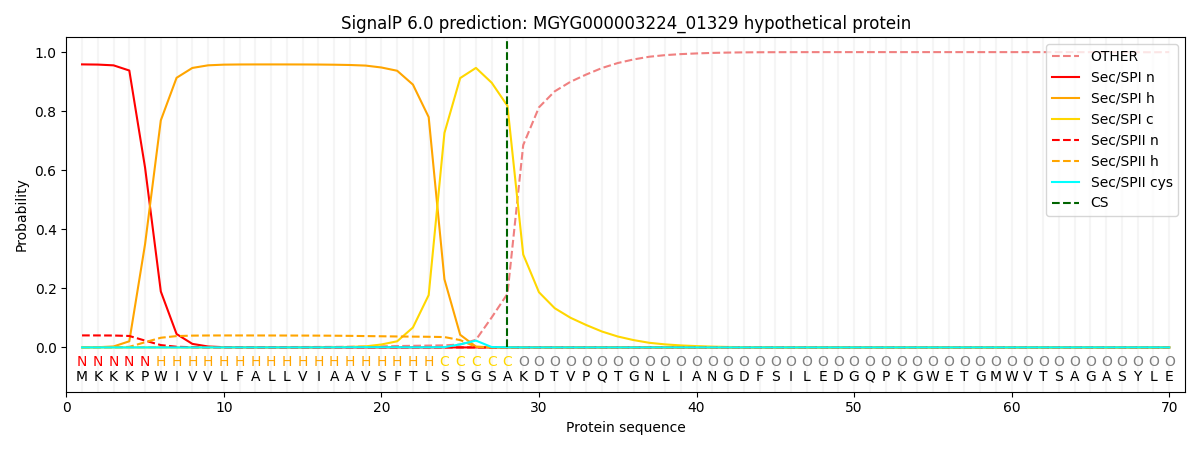
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.002131 | 0.954594 | 0.042496 | 0.000276 | 0.000247 | 0.000245 |
TMHMM Annotations download full data without filtering help
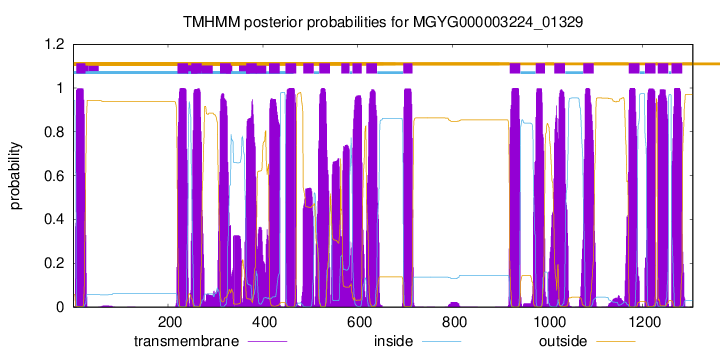
| start | end |
|---|---|
| 7 | 26 |
| 220 | 242 |
| 249 | 271 |
| 310 | 332 |
| 365 | 387 |
| 413 | 435 |
| 448 | 470 |
| 485 | 507 |
| 519 | 541 |
| 565 | 582 |
| 589 | 608 |
| 618 | 640 |
| 696 | 715 |
| 920 | 942 |
| 975 | 994 |
| 1014 | 1036 |
| 1075 | 1097 |
| 1171 | 1193 |
| 1205 | 1227 |
| 1232 | 1254 |
| 1261 | 1283 |
