You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003237_01624
You are here: Home > Sequence: MGYG000003237_01624
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Victivallis sp002998355 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; Victivallaceae; Victivallis; Victivallis sp002998355 | |||||||||||
| CAZyme ID | MGYG000003237_01624 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 19026; End: 21176 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 418 | 679 | 1.4e-32 | 0.6945454545454546 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 4.28e-22 | 416 | 676 | 32 | 272 | Cellulase (glycosyl hydrolase family 5). |
| pfam13472 | Lipase_GDSL_2 | 5.75e-11 | 41 | 232 | 3 | 175 | GDSL-like Lipase/Acylhydrolase family. This family of presumed lipases and related enzymes are similar to pfam00657. |
| cd00229 | SGNH_hydrolase | 4.03e-08 | 37 | 241 | 1 | 187 | SGNH_hydrolase, or GDSL_hydrolase, is a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the typical Ser-His-Asp(Glu) triad from other serine hydrolases, but may lack the carboxlic acid. |
| COG2730 | BglC | 1.25e-06 | 418 | 694 | 83 | 381 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| cd01834 | SGNH_hydrolase_like_2 | 1.00e-05 | 36 | 230 | 3 | 180 | SGNH_hydrolase subfamily. SGNH hydrolases are a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the Ser-His-Asp(Glu) triad found in other serine hydrolases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVM44090.1 | 0.0 | 1 | 716 | 1 | 716 |
| AVM45141.1 | 1.26e-88 | 258 | 714 | 169 | 634 |
| AVM43770.1 | 1.87e-86 | 249 | 687 | 647 | 1089 |
| AVV54443.1 | 5.56e-67 | 393 | 716 | 35 | 382 |
| AOH40543.1 | 5.56e-67 | 393 | 716 | 35 | 382 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3NCO_A | 2.95e-17 | 496 | 674 | 122 | 292 | Crystalstructure of FnCel5A from F. nodosum Rt17-B1 [Fervidobacterium nodosum Rt17-B1],3NCO_B Crystal structure of FnCel5A from F. nodosum Rt17-B1 [Fervidobacterium nodosum Rt17-B1] |
| 3RJX_A | 2.95e-17 | 496 | 674 | 122 | 292 | CrystalStructure of Hyperthermophilic Endo-Beta-1,4-glucanase [Fervidobacterium nodosum Rt17-B1] |
| 3RJY_A | 2.95e-17 | 496 | 674 | 122 | 292 | CrystalStructure of Hyperthermophilic Endo-beta-1,4-glucanase in complex with substrate [Fervidobacterium nodosum Rt17-B1] |
| 6KDD_A | 1.35e-16 | 496 | 684 | 122 | 306 | endoglucanase[Fervidobacterium pennivorans DSM 9078] |
| 3AMC_A | 9.91e-16 | 491 | 674 | 109 | 285 | Crystalstructures of Thermotoga maritima Cel5A, apo form and dimer/au [Thermotoga maritima MSB8],3AMC_B Crystal structures of Thermotoga maritima Cel5A, apo form and dimer/au [Thermotoga maritima MSB8],3AMD_A Crystal structures of Thermotoga maritima Cel5A, apo form and tetramer/au [Thermotoga maritima MSB8],3AMD_B Crystal structures of Thermotoga maritima Cel5A, apo form and tetramer/au [Thermotoga maritima MSB8],3AMD_C Crystal structures of Thermotoga maritima Cel5A, apo form and tetramer/au [Thermotoga maritima MSB8],3AMD_D Crystal structures of Thermotoga maritima Cel5A, apo form and tetramer/au [Thermotoga maritima MSB8],3MMU_A Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_B Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_C Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_D Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_E Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_F Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_G Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_H Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMW_A Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMW_B Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMW_C Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMW_D Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P16169 | 9.90e-07 | 438 | 671 | 62 | 287 | Cellodextrinase A OS=Ruminococcus flavefaciens OX=1265 GN=celA PE=3 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as SP
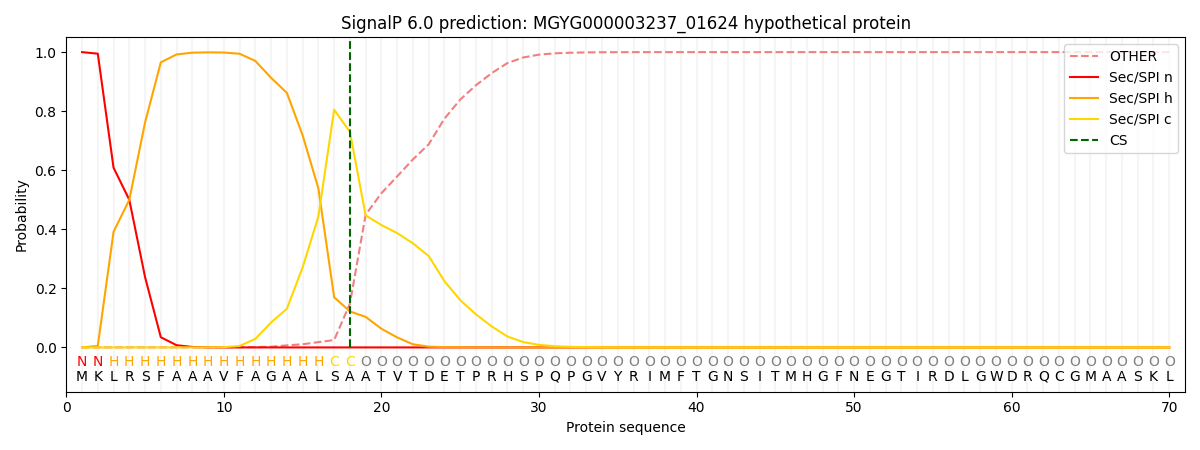
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000887 | 0.998084 | 0.000255 | 0.000275 | 0.000239 | 0.000208 |
