You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003237_01636
You are here: Home > Sequence: MGYG000003237_01636
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Victivallis sp002998355 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; Victivallaceae; Victivallis; Victivallis sp002998355 | |||||||||||
| CAZyme ID | MGYG000003237_01636 | |||||||||||
| CAZy Family | GH42 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 36833; End: 39226 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH42 | 202 | 440 | 4.8e-16 | 0.5795148247978437 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02449 | Glyco_hydro_42 | 9.15e-14 | 264 | 428 | 73 | 217 | Beta-galactosidase. This group of beta-galactosidase enzymes belong to the glycosyl hydrolase 42 family. The enzyme catalyzes the hydrolysis of terminal, non-reducing terminal beta-D-galactosidase residues. |
| COG1874 | GanA | 6.87e-10 | 172 | 444 | 9 | 254 | Beta-galactosidase GanA [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVM44099.1 | 0.0 | 1 | 797 | 1 | 797 |
| SDT33465.1 | 2.29e-53 | 167 | 680 | 51 | 570 |
| ASV74990.1 | 1.10e-36 | 235 | 649 | 267 | 659 |
| AQX14574.1 | 1.46e-08 | 178 | 441 | 5 | 242 |
| VEP38607.1 | 1.46e-08 | 178 | 441 | 5 | 242 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6Y2K_A | 9.77e-08 | 197 | 515 | 17 | 314 | ChainA, beta-galactosidase [Marinomonas sp. ef1] |
| 5E9A_A | 3.03e-07 | 267 | 540 | 120 | 388 | Crystalstructure analysis of the cold-adamped beta-galactosidase from Rahnella sp. R3 [Rahnella sp. R3],5E9A_B Crystal structure analysis of the cold-adamped beta-galactosidase from Rahnella sp. R3 [Rahnella sp. R3],5E9A_C Crystal structure analysis of the cold-adamped beta-galactosidase from Rahnella sp. R3 [Rahnella sp. R3],5E9A_D Crystal structure analysis of the cold-adamped beta-galactosidase from Rahnella sp. R3 [Rahnella sp. R3],5E9A_E Crystal structure analysis of the cold-adamped beta-galactosidase from Rahnella sp. R3 [Rahnella sp. R3],5E9A_F Crystal structure analysis of the cold-adamped beta-galactosidase from Rahnella sp. R3 [Rahnella sp. R3] |
| 4UCF_A | 5.16e-07 | 202 | 427 | 38 | 242 | Crystalstructure of Bifidobacterium bifidum beta-galactosidase in complex with alpha-galactose [Bifidobacterium bifidum S17],4UCF_B Crystal structure of Bifidobacterium bifidum beta-galactosidase in complex with alpha-galactose [Bifidobacterium bifidum S17],4UCF_C Crystal structure of Bifidobacterium bifidum beta-galactosidase in complex with alpha-galactose [Bifidobacterium bifidum S17],4UZS_A Crystal structure of Bifidobacterium bifidum beta-galactosidase [Bifidobacterium bifidum S17],4UZS_B Crystal structure of Bifidobacterium bifidum beta-galactosidase [Bifidobacterium bifidum S17],4UZS_C Crystal structure of Bifidobacterium bifidum beta-galactosidase [Bifidobacterium bifidum S17] |
| 1KWG_A | 3.41e-06 | 201 | 452 | 17 | 246 | Crystalstructure of Thermus thermophilus A4 beta-galactosidase [Thermus thermophilus],1KWK_A Crystal structure of Thermus thermophilus A4 beta-galactosidase in complex with galactose [Thermus thermophilus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8GEA9 | 1.58e-06 | 196 | 452 | 10 | 246 | Beta-galactosidase BgaA OS=Thermus sp. OX=275 GN=bgaA PE=1 SV=1 |
| D4QFE7 | 2.83e-06 | 202 | 427 | 38 | 242 | Beta-galactosidase BbgII OS=Bifidobacterium bifidum OX=1681 GN=bbgII PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
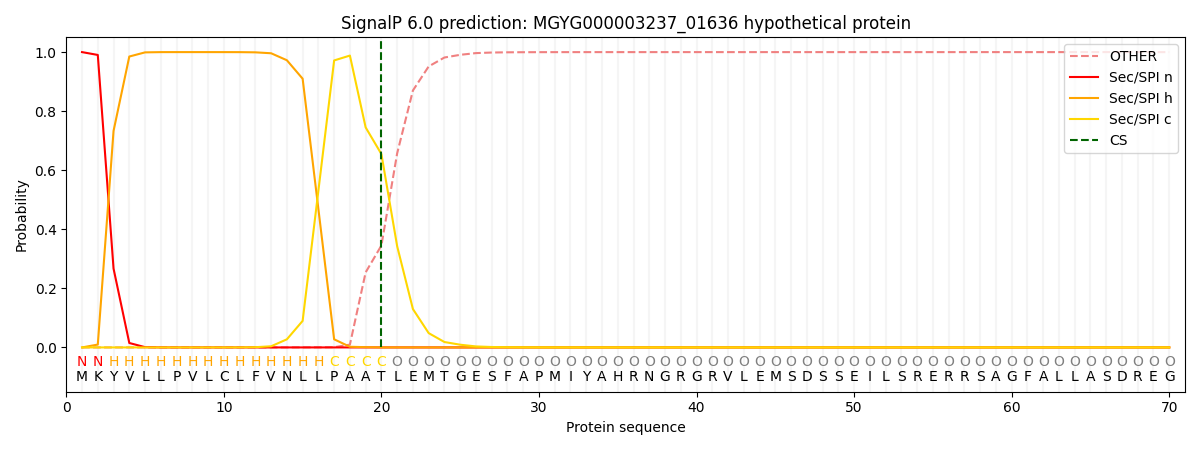
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000342 | 0.998947 | 0.000225 | 0.000153 | 0.000154 | 0.000160 |
