You are browsing environment: HUMAN GUT
MGYG000003237_02329
Basic Information
help
Species
Victivallis sp002998355
Lineage
Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; Victivallaceae; Victivallis; Victivallis sp002998355
CAZyme ID
MGYG000003237_02329
CAZy Family
GH39
CAZyme Description
hypothetical protein
CAZyme Property
Protein Length
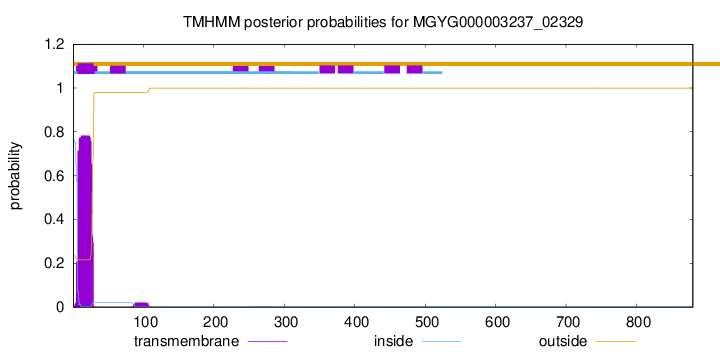
CGC
Molecular Weight
Isoelectric Point
880
97880.38
6.9692
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000003237
4618450
MAG
United States
North America
Gene Location
Start: 69555;
End: 72197
Strand: +
No EC number prediction in MGYG000003237_02329.
Family
Start
End
Evalue
family coverage
GH39
494
691
2.5e-16
0.4965197215777262
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
pfam17829
GH115_C
0.003
234
341
38
149
Gylcosyl hydrolase family 115 C-terminal domain. This domain is found at the C-terminus of glycosyl hydrolase family 115 proteins. This domain has a beta-sandwich fold.
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
AHF90237.1
6.72e-132
180
873
234
952
AFZ16313.1
7.39e-07
445
709
59
305
QJE96564.1
7.82e-06
11
237
6
221
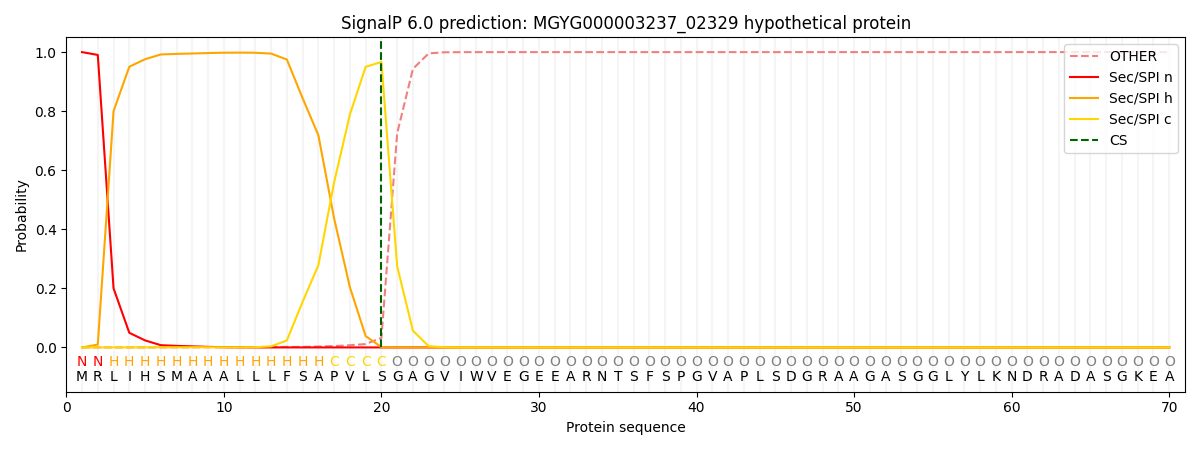
This protein is predicted as SP
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
0.000584
0.998466
0.000300
0.000225
0.000197
0.000194