You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003237_03711
You are here: Home > Sequence: MGYG000003237_03711
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Victivallis sp002998355 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; Victivallaceae; Victivallis; Victivallis sp002998355 | |||||||||||
| CAZyme ID | MGYG000003237_03711 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 89579; End: 93103 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 521 | 924 | 7.1e-35 | 0.43617021276595747 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 4.22e-17 | 522 | 944 | 76 | 497 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| pfam02836 | Glyco_hydro_2_C | 6.13e-08 | 724 | 941 | 3 | 209 | Glycosyl hydrolases family 2, TIM barrel domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
| pfam00703 | Glyco_hydro_2 | 1.20e-07 | 610 | 720 | 1 | 106 | Glycosyl hydrolases family 2. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
| PRK10340 | ebgA | 2.42e-06 | 610 | 850 | 209 | 449 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| PRK10150 | PRK10150 | 2.88e-06 | 669 | 889 | 235 | 446 | beta-D-glucuronidase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVM44993.1 | 0.0 | 1 | 1174 | 54 | 1227 |
| SEH75953.1 | 1.78e-19 | 514 | 942 | 264 | 668 |
| AHF94339.1 | 8.87e-17 | 596 | 952 | 211 | 538 |
| QDH71258.1 | 1.10e-16 | 514 | 965 | 81 | 526 |
| QKJ30744.1 | 1.33e-16 | 478 | 902 | 73 | 479 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5T98_A | 1.69e-11 | 681 | 889 | 256 | 448 | Crystalstructure of BuGH2Awt [Bacteroides uniformis],5T98_B Crystal structure of BuGH2Awt [Bacteroides uniformis],5T99_A Crystal structure of BuGH2Awt in complex with Galactoisofagomine [Bacteroides uniformis],5T99_B Crystal structure of BuGH2Awt in complex with Galactoisofagomine [Bacteroides uniformis] |
| 7CWD_A | 8.65e-11 | 622 | 893 | 182 | 438 | ChainA, beta-glalactosidase [Niallia circulans],7CWI_A Chain A, beta-galactosidase [Niallia circulans] |
| 6ED1_A | 3.62e-06 | 681 | 897 | 263 | 458 | ChainA, Glycosyl hydrolase family 2, sugar binding domain protein [Phocaeicola dorei],6ED1_B Chain B, Glycosyl hydrolase family 2, sugar binding domain protein [Phocaeicola dorei],6ED1_C Chain C, Glycosyl hydrolase family 2, sugar binding domain protein [Phocaeicola dorei],6ED1_D Chain D, Glycosyl hydrolase family 2, sugar binding domain protein [Phocaeicola dorei] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
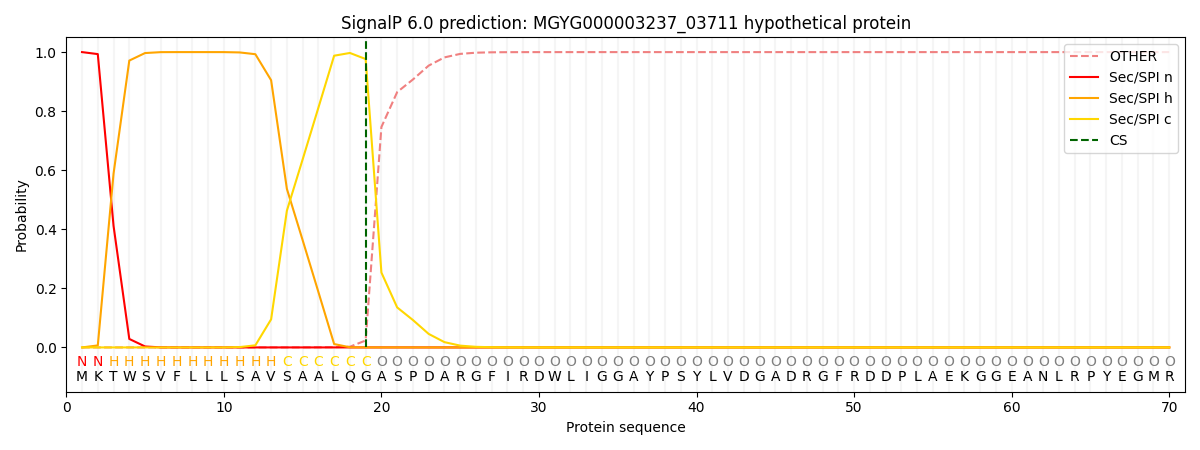
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000314 | 0.999029 | 0.000188 | 0.000158 | 0.000154 | 0.000150 |
