You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003238_00952
You are here: Home > Sequence: MGYG000003238_00952
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
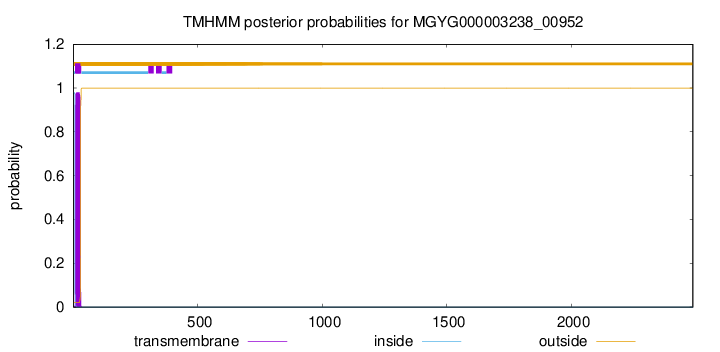
TMHMM annotations
Basic Information help
| Species | UBA1375 sp002305795 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; UBA1375; UBA1375 sp002305795 | |||||||||||
| CAZyme ID | MGYG000003238_00952 | |||||||||||
| CAZy Family | GH51 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 134; End: 7603 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH51 | 532 | 1313 | 2e-90 | 0.8793650793650793 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3534 | AbfA | 4.21e-32 | 602 | 985 | 45 | 388 | Alpha-L-arabinofuranosidase [Carbohydrate transport and metabolism]. |
| pfam06964 | Alpha-L-AF_C | 1.09e-15 | 906 | 988 | 20 | 93 | Alpha-L-arabinofuranosidase C-terminal domain. This family represents the C-terminus (approximately 200 residues) of bacterial and eukaryotic alpha-L-arabinofuranosidase (EC:3.2.1.55). This catalyzes the hydrolysis of nonreducing terminal alpha-L-arabinofuranosidic linkages in L-arabinose-containing polysaccharides. |
| smart00813 | Alpha-L-AF_C | 1.60e-15 | 906 | 983 | 16 | 85 | Alpha-L-arabinofuranosidase C-terminus. This entry represents the C terminus (approximately 200 residues) of bacterial and eukaryotic alpha-L-arabinofuranosidase. This catalyses the hydrolysis of non-reducing terminal alpha-L-arabinofuranosidic linkages in L-arabinose-containing polysaccharides. |
| COG5492 | YjdB | 6.16e-12 | 1556 | 1728 | 179 | 328 | Uncharacterized conserved protein YjdB, contains Ig-like domain [General function prediction only]. |
| pfam13385 | Laminin_G_3 | 1.29e-11 | 83 | 264 | 3 | 150 | Concanavalin A-like lectin/glucanases superfamily. This domain belongs to the Concanavalin A-like lectin/glucanases superfamily. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ANF95233.1 | 0.0 | 34 | 1316 | 4 | 1246 |
| AET58487.1 | 0.0 | 48 | 1314 | 23 | 1244 |
| AUO06232.1 | 0.0 | 48 | 1314 | 23 | 1244 |
| QOH60128.1 | 0.0 | 48 | 1314 | 23 | 1244 |
| AJE51460.1 | 0.0 | 48 | 1314 | 23 | 1244 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6ZPS_AAA | 1.28e-35 | 376 | 983 | 13 | 524 | ChainAAA, MgGH51 [Meripilus giganteus],6ZPV_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZPW_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZPX_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZPY_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZPZ_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZQ0_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZQ1_AAA Chain AAA, MgGH51 [Meripilus giganteus] |
| 2VRQ_A | 3.52e-11 | 607 | 1061 | 52 | 449 | StructureOf An Inactive Mutant Of Arabinofuranosidase From Thermobacillus Xylanilyticus In Complex With A Pentasaccharide [Thermobacillus xylanilyticus],2VRQ_B Structure Of An Inactive Mutant Of Arabinofuranosidase From Thermobacillus Xylanilyticus In Complex With A Pentasaccharide [Thermobacillus xylanilyticus],2VRQ_C Structure Of An Inactive Mutant Of Arabinofuranosidase From Thermobacillus Xylanilyticus In Complex With A Pentasaccharide [Thermobacillus xylanilyticus] |
| 4ATW_A | 4.45e-11 | 603 | 757 | 45 | 175 | Thecrystal structure of Arabinofuranosidase [Thermotoga maritima MSB8],4ATW_B The crystal structure of Arabinofuranosidase [Thermotoga maritima MSB8],4ATW_C The crystal structure of Arabinofuranosidase [Thermotoga maritima MSB8],4ATW_D The crystal structure of Arabinofuranosidase [Thermotoga maritima MSB8],4ATW_E The crystal structure of Arabinofuranosidase [Thermotoga maritima MSB8],4ATW_F The crystal structure of Arabinofuranosidase [Thermotoga maritima MSB8] |
| 3S2C_A | 4.48e-11 | 603 | 757 | 45 | 175 | Structureof the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_B Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_C Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_D Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_E Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_F Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_G Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_H Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_I Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_J Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_K Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_L Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1] |
| 6ZT6_A | 4.64e-11 | 607 | 1061 | 52 | 449 | ChainA, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT6_B Chain B, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT6_C Chain C, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT7_A Chain A, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT7_B Chain B, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT7_C Chain C, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P82593 | 1.84e-93 | 354 | 1313 | 26 | 823 | Extracellular exo-alpha-L-arabinofuranosidase OS=Streptomyces chartreusis OX=1969 PE=1 SV=1 |
| Q9SG80 | 7.54e-53 | 376 | 983 | 55 | 548 | Alpha-L-arabinofuranosidase 1 OS=Arabidopsis thaliana OX=3702 GN=ASD1 PE=1 SV=1 |
| Q8VZR2 | 1.81e-51 | 365 | 999 | 42 | 563 | Alpha-L-arabinofuranosidase 2 OS=Arabidopsis thaliana OX=3702 GN=ASD2 PE=2 SV=1 |
| Q96X54 | 5.30e-34 | 377 | 982 | 39 | 531 | Probable alpha-L-arabinofuranosidase A OS=Aspergillus awamori OX=105351 GN=abfA PE=1 SV=1 |
| Q8NK90 | 1.67e-33 | 377 | 982 | 39 | 531 | Alpha-L-arabinofuranosidase A OS=Aspergillus kawachii (strain NBRC 4308) OX=1033177 GN=abfA PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
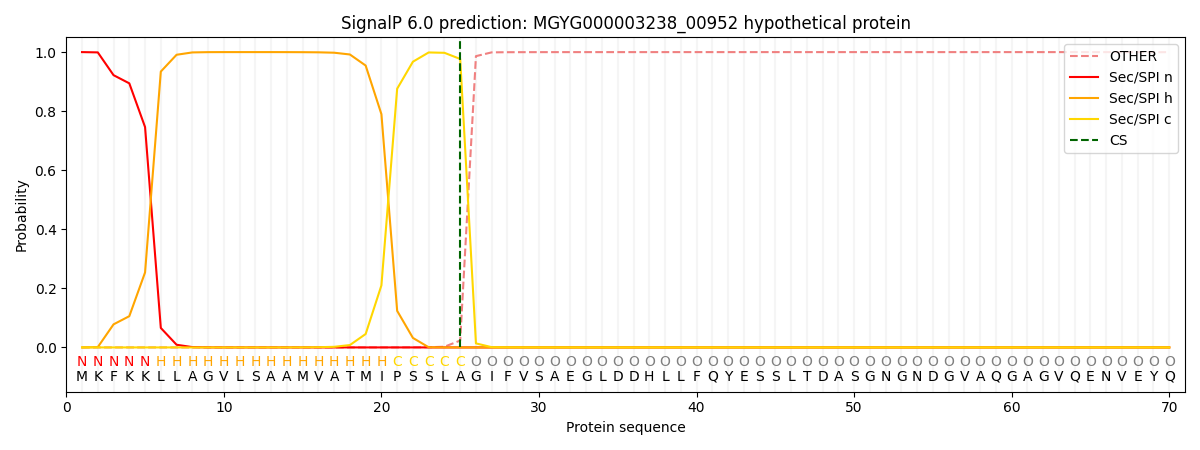
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000536 | 0.998477 | 0.000294 | 0.000245 | 0.000221 | 0.000205 |