You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003238_01178
You are here: Home > Sequence: MGYG000003238_01178
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
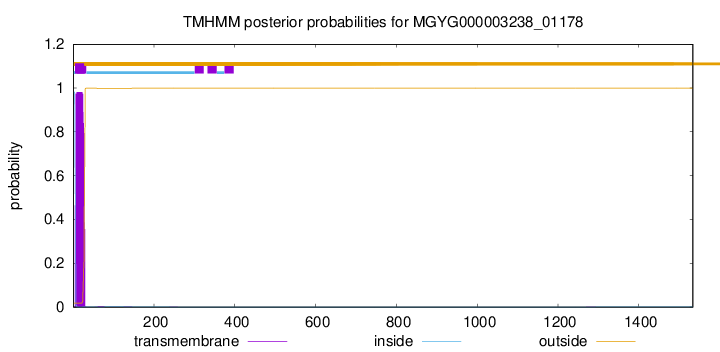
TMHMM annotations
Basic Information help
| Species | UBA1375 sp002305795 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; UBA1375; UBA1375 sp002305795 | |||||||||||
| CAZyme ID | MGYG000003238_01178 | |||||||||||
| CAZy Family | GH43 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 4166; End: 8770 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH43 | 1157 | 1446 | 1.7e-79 | 0.9966555183946488 |
| GH43 | 48 | 357 | 3e-70 | 0.9967741935483871 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd18819 | GH43_LbAraf43-like | 1.42e-113 | 1158 | 1416 | 1 | 277 | Glycosyl hydrolase family 43 proteins similar to Lactobacillus brevis alpha-L-arabinofuranosidase LbAraf43 and Geobacillus thermoleovorans GbtXyl43B. This uncharacterized glycosyl hydrolase family 43 (GH43) subgroup belongs to a subgroup which includes enzymes with beta-xylosidase (EC 3.2.1.37), alpha-L-arabinofuranosidase (EC 3.2.1.55) and possibly bifunctional xylosidase/arabinofuranosidase activities, similar to Lactobacillus brevis alpha-L-arabinofuranosidase LbAraf43 and Geobacillus thermoleovorans IT-08 beta-xylosidase / exo-xylanase (GbtXyl43B). It belongs to the glycosyl hydrolase clan F (according to carbohydrate-active enzymes database (CAZY)) which includes family 43 (GH43) and 62 (GH62) families. GH43 are inverting enzymes (i.e. they invert the stereochemistry of the anomeric carbon atom of the substrate) that have an aspartate as the catalytic general base, a glutamate as the catalytic general acid and another aspartate that is responsible for pKa modulation and orienting the catalytic acid. Many GH43 enzymes display both alpha-L-arabinofuranosidase and beta-D-xylosidase activity using aryl-glycosides as substrates. A common structural feature of GH43 enzymes is a 5-bladed beta-propeller domain that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
| cd18818 | GH43_GbtXyl43B-like | 1.40e-89 | 1158 | 1416 | 1 | 273 | Glycosyl hydrolase family 43 such as Geobacillus thermoleovorans IT-08 beta-xylosidase/exo-xylanase (GbtXyl43B). This glycosyl hydrolase family 43 (GH43) subgroup includes the characterized enzymes Geobacillus thermoleovorans IT-08 beta-xylosidase (EC 3.2.1.37) / exo-xylanase (GbtXyl43B), and Paenibacillus sp. strain E18 alpha-L-arabinofuranosidase (EC 3.2.1.55) Abf43B. It belongs to the glycosyl hydrolase clan F (according to carbohydrate-active enzymes database (CAZY)) which includes family 43 (GH43) and 62 (GH62) families. GH43 are inverting enzymes (i.e. they invert the stereochemistry of the anomeric carbon atom of the substrate) that have an aspartate as the catalytic general base, a glutamate as the catalytic general acid and another aspartate that is responsible for pKa modulation and orienting the catalytic acid. Many GH43 enzymes display both alpha-L-arabinofuranosidase and beta-D-xylosidase activity using aryl-glycosides as substrates. A common structural feature of GH43 enzymes is a 5-bladed beta-propeller domain that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
| cd18817 | GH43f_LbAraf43-like | 1.50e-86 | 1158 | 1416 | 1 | 262 | Glycosyl hydrolase family 43 such as Lactobacillus brevis alpha-L-arabinofuranosidase LbAraf43. This glycosyl hydrolase family 43 (GH43) subgroup includes characterized enzymes with alpha-L-arabinofuranosidase (EC 3.2.1.55) activity. It belongs to the glycosyl hydrolase clan F (according to carbohydrate-active enzymes database (CAZY)) which includes family 43 (GH43) and 62 (GH62) families. GH43 are inverting enzymes (i.e. they invert the stereochemistry of the anomeric carbon atom of the substrate) that have an aspartate as the catalytic general base, a glutamate as the catalytic general acid and another aspartate that is responsible for pKa modulation and orienting the catalytic acid. Many GH43 enzymes display both alpha-L-arabinofuranosidase and beta-D-xylosidase activity using aryl-glycosides as substrates. Characterized enzymes belonging to this subgroup include Lactobacillus brevis (LbAraf43) and Weissella sp (WAraf43) which show activity with similar catalytic efficiency on 1,5-alpha-L-arabinooligosaccharides with a degree of polymerization (DP) of 2-3; size is limited by an extended loop at the entrance to the active site. A common structural feature of GH43 enzymes is a 5-bladed beta-propeller domain that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
| COG3940 | COG3940 | 2.57e-85 | 1148 | 1446 | 4 | 308 | Beta-xylosidase, GH43 family [Carbohydrate transport and metabolism]. |
| cd08998 | GH43_Arb43a-like | 2.20e-75 | 49 | 352 | 1 | 278 | Glycosyl hydrolase family 43 protein such as Bacillus subtilis subsp. subtilis str. 168 endo-alpha-1,5-L-arabinanase Arb43A. This glycosyl hydrolase family 43 (GH43) subgroup belongs to the glycosyl hydrolase clan F (according to carbohydrate-active enzymes database (CAZY)) which includes family 43 (GH43) and 62 (GH62) families. GH43 are inverting enzymes (i.e. they invert the stereochemistry of the anomeric carbon atom of the substrate) that have an aspartate as the catalytic general base, a glutamate as the catalytic general acid and another aspartate that is responsible for pKa modulation and orienting the catalytic acid. The GH43 ABN enzymes hydrolyze alpha-1,5-L-arabinofuranoside linkages while the ABF enzymes cleave arabinose side chains so that the combined actions of these two enzymes reduce arabinan to L-arabinose and/or arabinooligosaccharides. Many of these enzymes such as the Bacillus subtilis arabinanase Abn2, that hydrolyzes sugar beet arabinan (branched), linear alpha-1,5-L-arabinan and pectin, are different from other arabinases; they are organized into two different domains with a divalent metal cluster close to the catalytic residues to guarantee the correct protonation state of the catalytic residues and consequently the enzyme activity. These arabinan-degrading enzymes are important in the food industry for efficient production of L-arabinose from agricultural waste; L-arabinose is often used as a bioactive sweetener. A common structural feature of GH43 enzymes is a 5-bladed beta-propeller domain that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBF42300.1 | 7.53e-176 | 506 | 1476 | 58 | 1067 |
| CAD6011226.1 | 2.55e-152 | 491 | 1469 | 22 | 1023 |
| QNF28727.1 | 3.26e-145 | 492 | 1471 | 36 | 1017 |
| AZZ51743.1 | 4.46e-131 | 494 | 1470 | 35 | 1037 |
| AIQ31573.1 | 1.70e-97 | 915 | 1469 | 651 | 1230 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3AKF_A | 3.25e-68 | 1149 | 1446 | 11 | 310 | Crystalstructure of exo-1,5-alpha-L-arabinofuranosidase [Streptomyces avermitilis MA-4680 = NBRC 14893],3AKG_A Crystal structure of exo-1,5-alpha-L-arabinofuranosidase complexed with alpha-1,5-L-arabinofuranobiose [Streptomyces avermitilis MA-4680 = NBRC 14893],3AKH_A Crystal structure of exo-1,5-alpha-L-arabinofuranosidase complexed with alpha-1,5-L-arabinofuranotriose [Streptomyces avermitilis MA-4680 = NBRC 14893],3AKI_A Crystal structure of exo-1,5-alpha-L-arabinofuranosidase complexed with alpha-L-arabinofuranosyl azido [Streptomyces avermitilis MA-4680 = NBRC 14893] |
| 3K1U_A | 1.05e-66 | 1145 | 1459 | 5 | 325 | Beta-xylosidase,family 43 glycosyl hydrolase from Clostridium acetobutylicum [Clostridium acetobutylicum] |
| 5M8B_A | 2.71e-63 | 1144 | 1446 | 22 | 335 | ChainA, Alpha-L-arabinofuranosidase II [Levilactobacillus brevis],5M8B_B Chain B, Alpha-L-arabinofuranosidase II [Levilactobacillus brevis] |
| 5M8E_A | 6.44e-61 | 1142 | 1447 | 18 | 334 | Crystalstructure of a GH43 arabonofuranosidase from Weissella sp. strain 142 [Weissella cibaria],5M8E_B Crystal structure of a GH43 arabonofuranosidase from Weissella sp. strain 142 [Weissella cibaria] |
| 4KC7_A | 1.33e-34 | 47 | 419 | 31 | 419 | CrystalStructure of Endo-1,5-alpha-L-arabinanase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],4KC7_B Crystal Structure of Endo-1,5-alpha-L-arabinanase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],4KC7_C Crystal Structure of Endo-1,5-alpha-L-arabinanase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],4KC8_A Crystal Structure of Endo-1,5-alpha-L-arabinanase from Thermotoga petrophila RKU-1 in complex with TRIS [Thermotoga petrophila RKU-1],4KC8_B Crystal Structure of Endo-1,5-alpha-L-arabinanase from Thermotoga petrophila RKU-1 in complex with TRIS [Thermotoga petrophila RKU-1],4KC8_C Crystal Structure of Endo-1,5-alpha-L-arabinanase from Thermotoga petrophila RKU-1 in complex with TRIS [Thermotoga petrophila RKU-1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q82P90 | 2.49e-67 | 1149 | 1446 | 37 | 336 | Extracellular exo-alpha-(1->5)-L-arabinofuranosidase OS=Streptomyces avermitilis (strain ATCC 31267 / DSM 46492 / JCM 5070 / NBRC 14893 / NCIMB 12804 / NRRL 8165 / MA-4680) OX=227882 GN=Araf43A PE=1 SV=1 |
| P82594 | 4.81e-57 | 1149 | 1424 | 50 | 327 | Extracellular exo-alpha-(1->5)-L-arabinofuranosidase OS=Streptomyces chartreusis OX=1969 PE=1 SV=1 |
| G4MMH2 | 5.60e-57 | 1149 | 1446 | 41 | 342 | Alpha-L-arabinofuranosidase B OS=Magnaporthe oryzae (strain 70-15 / ATCC MYA-4617 / FGSC 8958) OX=242507 GN=abfB PE=1 SV=1 |
| A5IKD4 | 6.93e-34 | 47 | 419 | 28 | 416 | Extracellular endo-alpha-(1->5)-L-arabinanase OS=Thermotoga petrophila (strain ATCC BAA-488 / DSM 13995 / JCM 10881 / RKU-1) OX=390874 GN=Tpet_0637 PE=1 SV=1 |
| P42293 | 1.22e-33 | 46 | 426 | 33 | 427 | Extracellular endo-alpha-(1->5)-L-arabinanase 2 OS=Bacillus subtilis (strain 168) OX=224308 GN=abn2 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
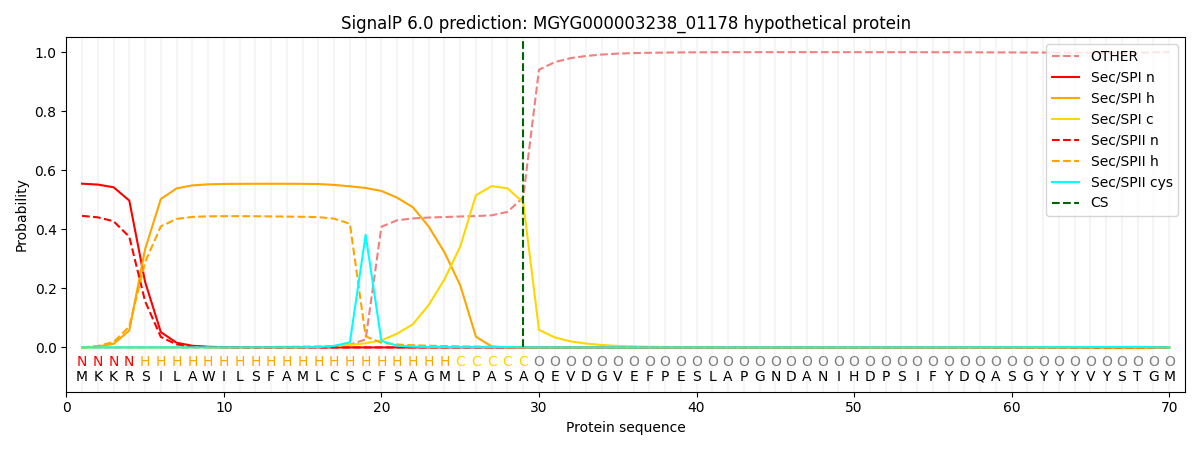
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000530 | 0.544685 | 0.453893 | 0.000374 | 0.000276 | 0.000217 |