You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003249_01721
You are here: Home > Sequence: MGYG000003249_01721
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Victivallis sp900761715 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; Victivallaceae; Victivallis; Victivallis sp900761715 | |||||||||||
| CAZyme ID | MGYG000003249_01721 | |||||||||||
| CAZy Family | GH110 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 193; End: 2250 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH110 | 114 | 616 | 5.1e-42 | 0.9197080291970803 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13229 | Beta_helix | 1.46e-08 | 474 | 615 | 1 | 112 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam13229 | Beta_helix | 1.18e-06 | 480 | 611 | 52 | 156 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVM46891.1 | 0.0 | 1 | 685 | 1 | 685 |
| SDT96813.1 | 4.16e-86 | 117 | 657 | 2 | 556 |
| CCW36343.1 | 4.64e-67 | 97 | 677 | 45 | 613 |
| SDS15045.1 | 5.47e-42 | 115 | 676 | 63 | 622 |
| AHF90434.1 | 7.13e-42 | 112 | 645 | 26 | 571 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q89ZX0 | 1.14e-11 | 117 | 633 | 47 | 573 | Alpha-1,3-galactosidase B OS=Bacteroides thetaiotaomicron (strain ATCC 29148 / DSM 2079 / JCM 5827 / CCUG 10774 / NCTC 10582 / VPI-5482 / E50) OX=226186 GN=glaB PE=3 SV=1 |
| B1KD83 | 3.13e-10 | 133 | 656 | 80 | 604 | Alpha-1,3-galactosidase A OS=Shewanella woodyi (strain ATCC 51908 / MS32) OX=392500 GN=glaA PE=3 SV=1 |
| B2UNU8 | 3.34e-09 | 117 | 611 | 238 | 738 | Alpha-1,3-galactosidase B OS=Akkermansia muciniphila (strain ATCC BAA-835 / DSM 22959 / JCM 33894 / BCRC 81048 / CCUG 64013 / CIP 107961 / Muc) OX=349741 GN=glaB PE=3 SV=1 |
| A6L2M8 | 3.12e-07 | 368 | 612 | 317 | 563 | Alpha-1,3-galactosidase B OS=Phocaeicola vulgatus (strain ATCC 8482 / DSM 1447 / JCM 5826 / CCUG 4940 / NBRC 14291 / NCTC 11154) OX=435590 GN=glaB2 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
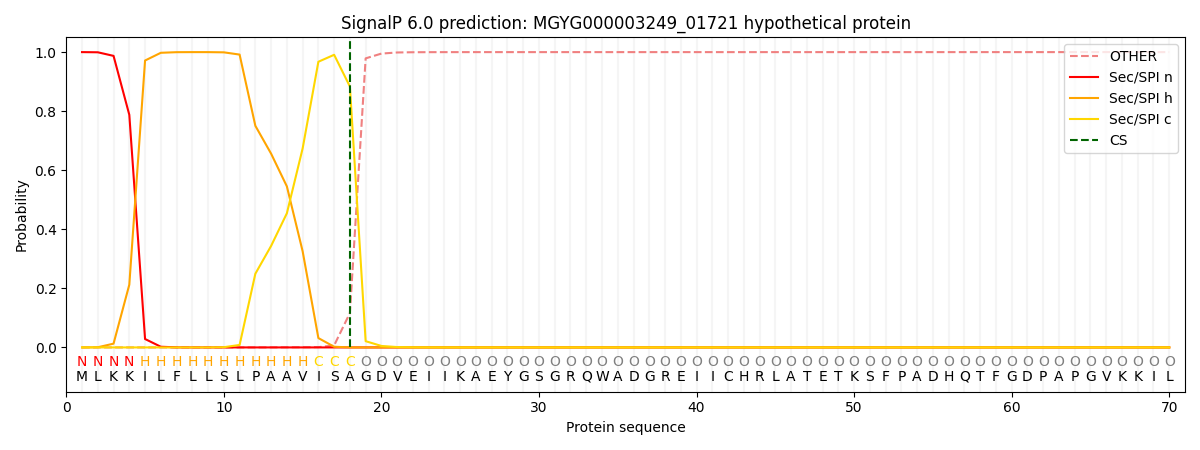
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000267 | 0.999073 | 0.000161 | 0.000163 | 0.000155 | 0.000145 |
