You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003249_02443
You are here: Home > Sequence: MGYG000003249_02443
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Victivallis sp900761715 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; Victivallaceae; Victivallis; Victivallis sp900761715 | |||||||||||
| CAZyme ID | MGYG000003249_02443 | |||||||||||
| CAZy Family | GH50 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2033; End: 4009 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH50 | 22 | 491 | 4.4e-85 | 0.7718223583460949 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVM46385.1 | 2.02e-138 | 6 | 657 | 8 | 669 |
| QRO01254.1 | 1.99e-100 | 18 | 515 | 213 | 712 |
| BBI31951.1 | 1.03e-82 | 22 | 496 | 610 | 1102 |
| CCW35847.1 | 3.60e-66 | 22 | 496 | 198 | 695 |
| QBG46742.1 | 3.20e-61 | 22 | 497 | 231 | 749 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6XJ9_A | 4.38e-49 | 20 | 492 | 221 | 760 | Structureof PfGH50B [Pseudoalteromonas fuliginea],6XJ9_B Structure of PfGH50B [Pseudoalteromonas fuliginea] |
| 5Z6P_A | 5.95e-49 | 16 | 491 | 213 | 759 | Thecrystal structure of an agarase, AgWH50C [Agarivorans gilvus],5Z6P_B The crystal structure of an agarase, AgWH50C [Agarivorans gilvus] |
| 4BQ2_A | 1.31e-48 | 22 | 491 | 204 | 743 | Structuralanalysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ2_B Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ2_C Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ2_D Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ3_A Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ3_B Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ3_C Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ3_D Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40] |
| 4BQ4_A | 3.29e-48 | 22 | 491 | 204 | 743 | Structuralanalysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ4_B Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ5_A Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ5_B Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P48840 | 9.35e-54 | 22 | 491 | 432 | 949 | Beta-agarase B OS=Vibrio sp. (strain JT0107) OX=47913 GN=agaB PE=3 SV=1 |
| P48839 | 6.51e-26 | 23 | 503 | 400 | 925 | Beta-agarase A OS=Vibrio sp. (strain JT0107) OX=47913 GN=agaA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
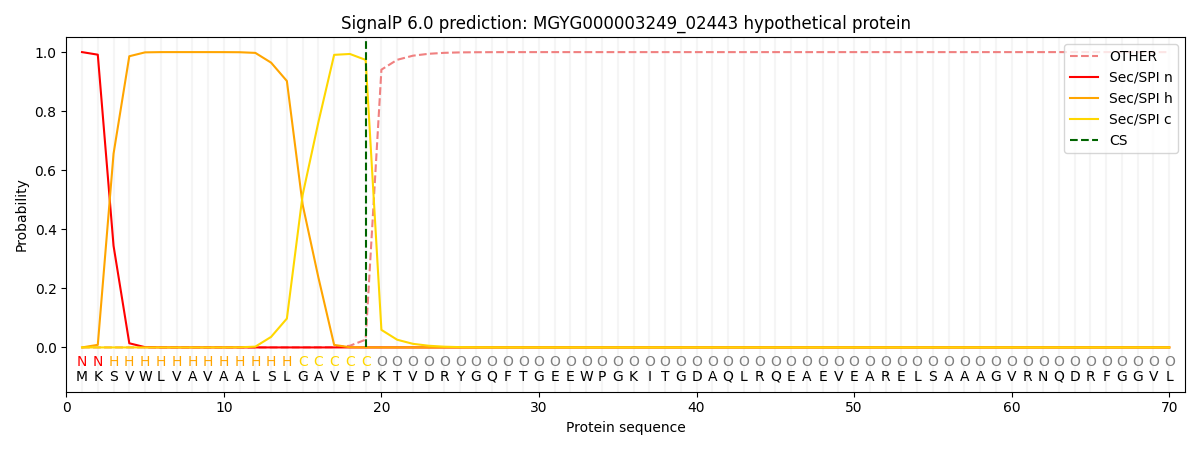
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000392 | 0.998823 | 0.000184 | 0.000204 | 0.000190 | 0.000178 |
