You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003252_02248
You are here: Home > Sequence: MGYG000003252_02248
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides sp900761785 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides sp900761785 | |||||||||||
| CAZyme ID | MGYG000003252_02248 | |||||||||||
| CAZy Family | GH33 | |||||||||||
| CAZyme Description | Sialidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 19539; End: 20804 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH33 | 57 | 397 | 5.6e-60 | 0.9444444444444444 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd15482 | Sialidase_non-viral | 2.00e-63 | 58 | 404 | 2 | 334 | Non-viral sialidases. Sialidases or neuraminidases function to bind and hydrolyze terminal sialic acid residues from various glycoconjugates, they play vital roles in pathogenesis, bacterial nutrition and cellular interactions. They have a six-bladed, beta-propeller fold with the non-viral sialidases containing 2-5 Asp-box motifs (most commonly Ser/Thr-X-Asp-[X]-Gly-X-Thr- Trp/Phe). This CD includes eubacterial and eukaryotic sialidases. |
| COG4409 | NanH | 1.26e-14 | 34 | 305 | 236 | 547 | Neuraminidase (sialidase) [Carbohydrate transport and metabolism, Cell wall/membrane/envelope biogenesis]. |
| pfam13088 | BNR_2 | 2.85e-12 | 81 | 318 | 1 | 215 | BNR repeat-like domain. This family of proteins contains BNR-like repeats suggesting these proteins may act as sialidases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUR43337.1 | 1.47e-305 | 1 | 421 | 1 | 425 |
| QIU95629.1 | 1.12e-288 | 1 | 421 | 1 | 423 |
| QIU95628.1 | 1.13e-77 | 55 | 418 | 35 | 382 |
| QUR43338.1 | 2.16e-74 | 55 | 418 | 35 | 382 |
| SEH77633.1 | 6.05e-66 | 58 | 417 | 69 | 403 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7MHU_A | 5.40e-286 | 22 | 421 | 2 | 401 | ChainA, Exo-alpha-sialidase [Bacteroides acidifaciens],7MHU_B Chain B, Exo-alpha-sialidase [Bacteroides acidifaciens] |
| 7LBU_A | 3.71e-22 | 55 | 411 | 79 | 427 | ChainA, Exo-alpha-sialidase [Cutibacterium acnes],7LBV_A Chain A, Exo-alpha-sialidase [Cutibacterium acnes] |
| 3SIL_A | 4.01e-21 | 58 | 404 | 20 | 369 | ChainA, SIALIDASE [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],7AEY_AAA Chain AAA, Sialidase [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2] |
| 1DIL_A | 4.09e-21 | 58 | 404 | 22 | 371 | SialidaseFrom Salmonella Typhimurium Complexed With Apana And Epana Inhibitors [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],1DIM_A Sialidase From Salmonella Typhimurium Complexed With Epana Inhibitor [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],2SIL_A Chain A, SIALIDASE [Salmonella enterica subsp. enterica serovar Typhimurium],2SIM_A Chain A, Sialidase [Salmonella enterica subsp. enterica serovar Typhimurium] |
| 6TRG_XXX | 2.48e-20 | 58 | 404 | 20 | 369 | ChainXXX, Sialidase [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],6TVI_AAA Chain AAA, Sialidase [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P29768 | 2.26e-20 | 58 | 404 | 23 | 372 | Sialidase OS=Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) OX=99287 GN=nanH PE=1 SV=4 |
| Q02834 | 1.01e-18 | 69 | 317 | 66 | 313 | Sialidase OS=Micromonospora viridifaciens OX=1881 GN=nedA PE=1 SV=1 |
| A5PF10 | 2.46e-15 | 69 | 383 | 77 | 377 | Sialidase-1 OS=Sus scrofa OX=9823 GN=NEU1 PE=3 SV=1 |
| Q5RAF4 | 3.27e-15 | 69 | 388 | 76 | 380 | Sialidase-1 OS=Pongo abelii OX=9601 GN=NEU1 PE=2 SV=1 |
| Q99519 | 3.27e-15 | 69 | 388 | 76 | 380 | Sialidase-1 OS=Homo sapiens OX=9606 GN=NEU1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
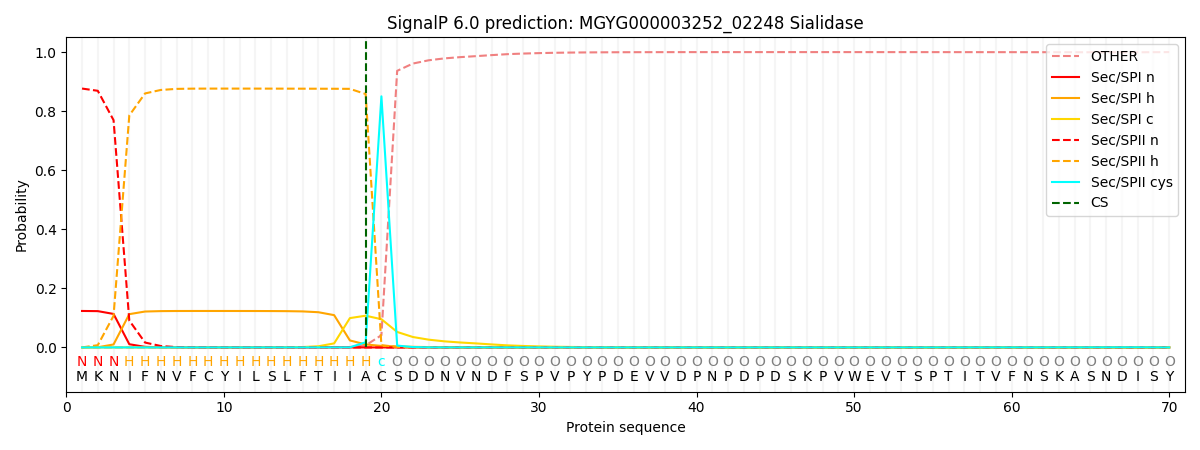
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000211 | 0.120478 | 0.879159 | 0.000056 | 0.000068 | 0.000059 |
