You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003252_03269
You are here: Home > Sequence: MGYG000003252_03269
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides sp900761785 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides sp900761785 | |||||||||||
| CAZyme ID | MGYG000003252_03269 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 62035; End: 64446 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13149 | Mfa_like_1 | 1.64e-28 | 49 | 283 | 1 | 231 | Fimbrillin-like. A family of putative fimbrillin proteins found by clustering human gut metagenomic sequences. Analysis of structural comparisons shows this family to be part of the FimbA (CL0450) superfamily of adhesin components or fimbrillins. |
| PLN02682 | PLN02682 | 9.37e-24 | 470 | 707 | 67 | 284 | pectinesterase family protein |
| COG4677 | PemB | 1.88e-21 | 463 | 740 | 72 | 362 | Pectin methylesterase and related acyl-CoA thioesterases [Carbohydrate transport and metabolism, Lipid transport and metabolism]. |
| PLN02432 | PLN02432 | 5.83e-21 | 482 | 701 | 20 | 208 | putative pectinesterase |
| PLN02773 | PLN02773 | 1.72e-20 | 480 | 699 | 12 | 213 | pectinesterase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QDH54500.1 | 0.0 | 1 | 803 | 1 | 803 |
| QIU94514.1 | 0.0 | 1 | 803 | 1 | 804 |
| QNL41098.1 | 0.0 | 1 | 803 | 1 | 803 |
| QDM11687.1 | 0.0 | 1 | 803 | 1 | 803 |
| QUT78059.1 | 0.0 | 1 | 803 | 1 | 803 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3UW0_A | 6.38e-13 | 466 | 723 | 25 | 282 | Pectinmethylesterase from Yersinia enterocolitica [Yersinia enterocolitica subsp. enterocolitica 8081] |
| 1GQ8_A | 6.14e-10 | 466 | 668 | 3 | 174 | Pectinmethylesterase from Carrot [Daucus carota] |
| 5C1C_A | 5.31e-09 | 583 | 711 | 98 | 222 | CrystalStructure of the Pectin Methylesterase from Aspergillus niger in Deglycosylated Form [Aspergillus niger ATCC 1015] |
| 5C1E_A | 5.31e-09 | 583 | 711 | 98 | 222 | CrystalStructure of the Pectin Methylesterase from Aspergillus niger in Penultimately Deglycosylated Form (N-acetylglucosamine Stub at Asn84) [Aspergillus niger ATCC 1015] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9LVQ0 | 1.15e-14 | 480 | 695 | 12 | 210 | Pectinesterase 31 OS=Arabidopsis thaliana OX=3702 GN=PME31 PE=1 SV=1 |
| Q9SIJ9 | 9.77e-14 | 484 | 701 | 60 | 248 | Putative pectinesterase 11 OS=Arabidopsis thaliana OX=3702 GN=PME11 PE=3 SV=1 |
| Q8VYZ3 | 1.70e-13 | 462 | 700 | 73 | 296 | Probable pectinesterase 53 OS=Arabidopsis thaliana OX=3702 GN=PME53 PE=2 SV=1 |
| D8VPP5 | 1.47e-12 | 590 | 697 | 158 | 270 | Pectinesterase 1 OS=Olea europaea OX=4146 PE=1 SV=1 |
| B2VPR8 | 1.47e-12 | 590 | 697 | 158 | 270 | Pectinesterase 2 OS=Olea europaea OX=4146 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
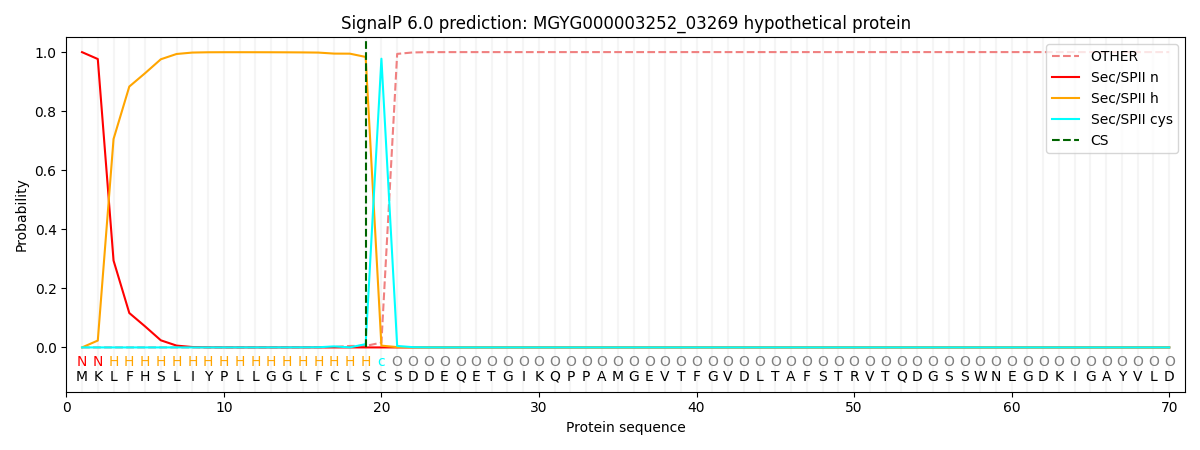
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000003 | 0.000188 | 0.999853 | 0.000000 | 0.000000 | 0.000000 |
