You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003252_03688
You are here: Home > Sequence: MGYG000003252_03688
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides sp900761785 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides sp900761785 | |||||||||||
| CAZyme ID | MGYG000003252_03688 | |||||||||||
| CAZy Family | GH28 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 9339; End: 10787 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH28 | 194 | 450 | 2.9e-36 | 0.7630769230769231 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PLN02218 | PLN02218 | 1.63e-07 | 232 | 452 | 206 | 414 | polygalacturonase ADPG |
| COG5434 | Pgu1 | 6.75e-06 | 205 | 382 | 225 | 392 | Polygalacturonase [Carbohydrate transport and metabolism]. |
| pfam00295 | Glyco_hydro_28 | 1.68e-05 | 267 | 453 | 139 | 310 | Glycosyl hydrolases family 28. Glycosyl hydrolase family 28 includes polygalacturonase EC:3.2.1.15 as well as rhamnogalacturonase A(RGase A), EC:3.2.1.-. These enzymes are important in cell wall metabolism. |
| PLN03003 | PLN03003 | 4.49e-04 | 204 | 390 | 118 | 298 | Probable polygalacturonase At3g15720 |
| PLN03010 | PLN03010 | 7.37e-04 | 184 | 460 | 118 | 383 | polygalacturonase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AAO77485.1 | 0.0 | 1 | 482 | 1 | 482 |
| QQA09361.1 | 0.0 | 1 | 482 | 1 | 482 |
| QMW84829.1 | 0.0 | 1 | 482 | 1 | 482 |
| BCA51661.1 | 0.0 | 1 | 482 | 1 | 482 |
| QUT72641.1 | 0.0 | 1 | 482 | 1 | 482 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A1DBR6 | 6.40e-08 | 173 | 417 | 162 | 422 | Probable endopolygalacturonase D OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=pgaD PE=3 SV=1 |
| Q5AYH4 | 2.10e-07 | 163 | 439 | 212 | 496 | Probable endopolygalacturonase D OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=pgaD PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
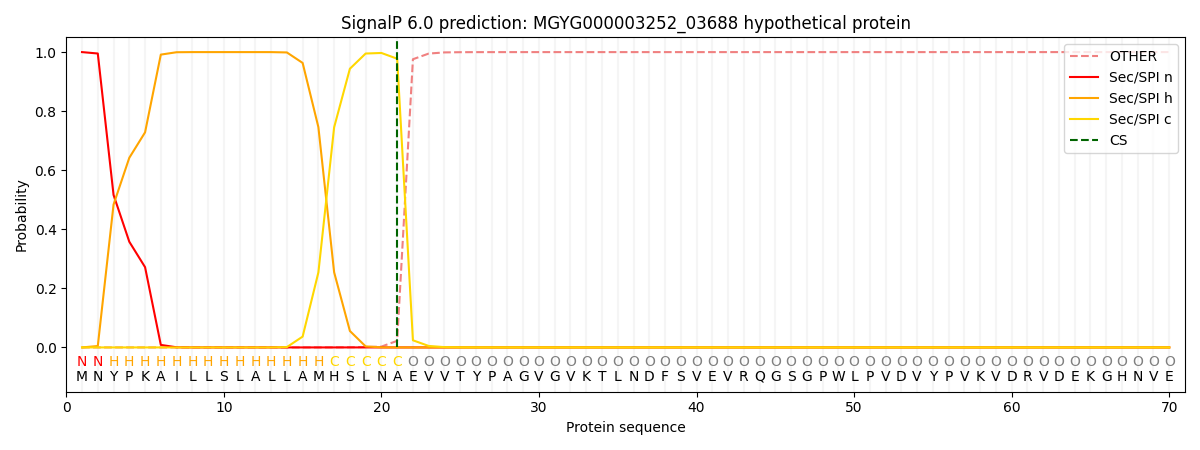
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000231 | 0.999147 | 0.000158 | 0.000153 | 0.000144 | 0.000135 |
