You are browsing environment: HUMAN GUT
MGYG000003252_04552
Basic Information
help
Species
Bacteroides sp900761785
Lineage
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides sp900761785
CAZyme ID
MGYG000003252_04552
CAZy Family
GT0
CAZyme Description
hypothetical protein
CAZyme Property
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000003252
8296673
MAG
United States
North America
Gene Location
Start: 12445;
End: 13389
Strand: +
No EC number prediction in MGYG000003252_04552.
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
pfam13641
Glyco_tranf_2_3
0.002
85
206
88
215
Glycosyltransferase like family 2. Members of this family of prokaryotic proteins include putative glucosyltransferase, which are involved in bacterial capsule biosynthesis.
more
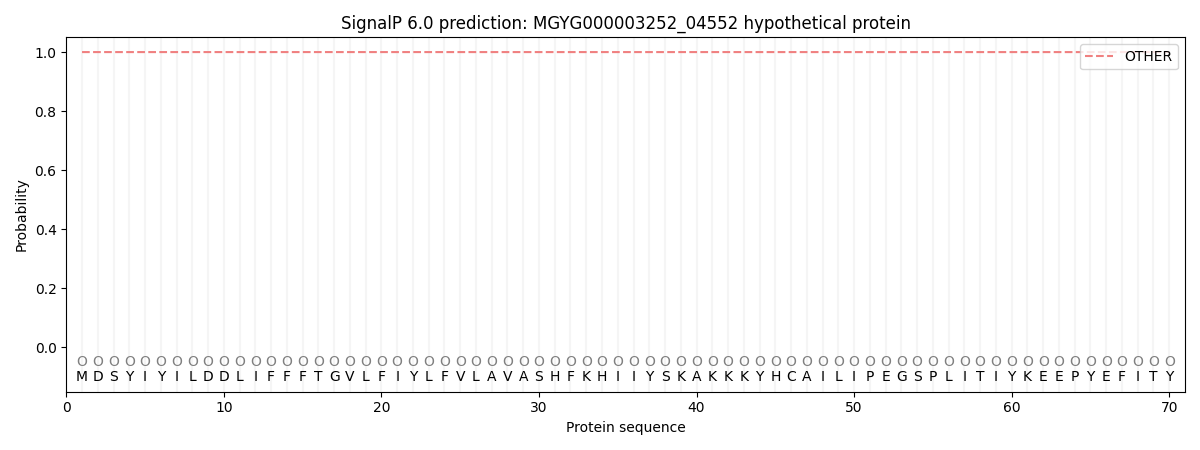
This protein is predicted as OTHER
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
1.000060
0.000006
0.000000
0.000000
0.000000
0.000000
start
end
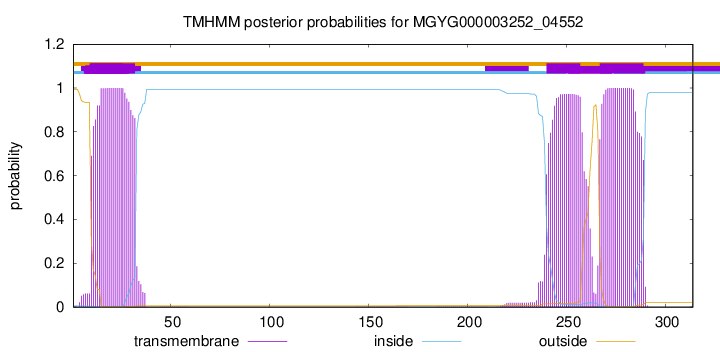
10
32
240
257
267
289