You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003267_00356
You are here: Home > Sequence: MGYG000003267_00356
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Xanthomonadales; Xanthomonadaceae; Stenotrophomonas; | |||||||||||
| CAZyme ID | MGYG000003267_00356 | |||||||||||
| CAZy Family | GT83 | |||||||||||
| CAZyme Description | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1312; End: 3042 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT83 | 6 | 519 | 2.1e-96 | 0.9388888888888889 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1807 | ArnT | 1.11e-39 | 1 | 532 | 1 | 517 | 4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| pfam13231 | PMT_2 | 3.52e-10 | 63 | 182 | 1 | 119 | Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366. |
| PRK13279 | arnT | 2.02e-06 | 8 | 179 | 5 | 181 | lipid IV(A) 4-amino-4-deoxy-L-arabinosyltransferase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AHY57527.1 | 7.01e-279 | 10 | 569 | 10 | 569 |
| AWH18344.1 | 1.90e-275 | 5 | 567 | 5 | 567 |
| AWH22891.1 | 5.42e-275 | 5 | 567 | 5 | 567 |
| AWH50959.1 | 1.86e-273 | 5 | 567 | 5 | 567 |
| AWH34584.1 | 2.55e-273 | 5 | 567 | 5 | 567 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5EZM_A | 1.09e-12 | 8 | 335 | 31 | 356 | CrystalStructure of ArnT from Cupriavidus metallidurans in the apo state [Cupriavidus metallidurans CH34],5F15_A Crystal Structure of ArnT from Cupriavidus metallidurans bound to Undecaprenyl phosphate [Cupriavidus metallidurans CH34] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q4ZSZ0 | 3.48e-22 | 12 | 340 | 11 | 329 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Pseudomonas syringae pv. syringae (strain B728a) OX=205918 GN=arnT PE=3 SV=1 |
| C6DAW3 | 3.55e-21 | 13 | 346 | 13 | 338 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Pectobacterium carotovorum subsp. carotovorum (strain PC1) OX=561230 GN=arnT PE=3 SV=1 |
| Q6D2F3 | 1.97e-20 | 18 | 346 | 18 | 338 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Pectobacterium atrosepticum (strain SCRI 1043 / ATCC BAA-672) OX=218491 GN=arnT PE=3 SV=1 |
| Q4KC80 | 7.83e-19 | 8 | 337 | 2 | 325 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase 1 OS=Pseudomonas fluorescens (strain ATCC BAA-477 / NRRL B-23932 / Pf-5) OX=220664 GN=arnT1 PE=3 SV=1 |
| Q2NRV9 | 1.39e-18 | 13 | 340 | 10 | 329 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase 2 OS=Sodalis glossinidius (strain morsitans) OX=343509 GN=arnT2 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000031 | 0.000014 | 0.000003 | 0.000000 | 0.000000 | 0.000004 |
TMHMM Annotations download full data without filtering help
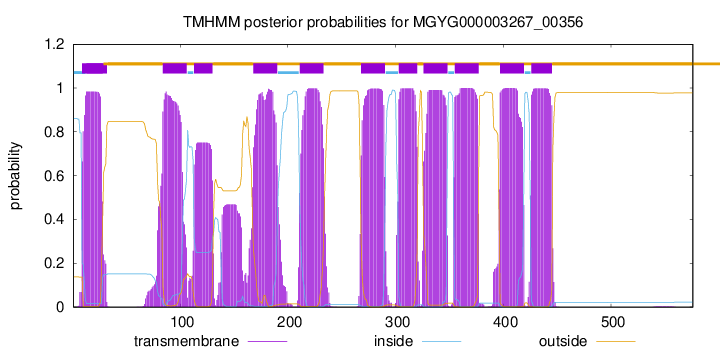
| start | end |
|---|---|
| 9 | 28 |
| 84 | 106 |
| 113 | 130 |
| 168 | 190 |
| 211 | 233 |
| 268 | 290 |
| 303 | 320 |
| 326 | 348 |
| 355 | 377 |
| 397 | 419 |
| 426 | 445 |
