You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003277_00736
You are here: Home > Sequence: MGYG000003277_00736
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Duncaniella sp900762315 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; Duncaniella; Duncaniella sp900762315 | |||||||||||
| CAZyme ID | MGYG000003277_00736 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 10889; End: 15124 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 46 | 458 | 1.3e-104 | 0.48404255319148937 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 8.15e-42 | 63 | 443 | 15 | 406 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK10150 | PRK10150 | 1.26e-32 | 62 | 443 | 14 | 417 | beta-D-glucuronidase; Provisional |
| PRK10340 | ebgA | 1.39e-23 | 60 | 446 | 41 | 453 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| pfam02837 | Glyco_hydro_2_N | 9.95e-16 | 21 | 212 | 3 | 165 | Glycosyl hydrolases family 2, sugar binding domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities and has a jelly-roll fold. The domain binds the sugar moiety during the sugar-hydrolysis reaction. |
| pfam13517 | VCBS | 5.09e-12 | 1009 | 1071 | 1 | 61 | Repeat domain in Vibrio, Colwellia, Bradyrhizobium and Shewanella. This domain of about 100 residues is found in multiple (up to 35) copies in long proteins from several species of Vibrio, Colwellia, Bradyrhizobium, and Shewanella (hence the name VCBS) and in smaller copy numbers in proteins from several other bacteria. The large protein size and repeat copy numbers, species distribution, and suggested activities of several member proteins suggests a role for this domain in adhesion (TIGR). |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT74091.1 | 0.0 | 21 | 1394 | 20 | 1415 |
| AGA25352.1 | 3.68e-214 | 23 | 768 | 24 | 755 |
| AQQ08666.1 | 4.52e-214 | 2 | 771 | 3 | 745 |
| QEM14171.1 | 1.89e-212 | 18 | 764 | 15 | 736 |
| ARN55733.1 | 5.38e-212 | 2 | 767 | 3 | 742 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7SF2_A | 6.65e-156 | 25 | 593 | 4 | 575 | ChainA, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_B Chain B, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_C Chain C, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_D Chain D, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_E Chain E, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_F Chain F, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838] |
| 3GM8_A | 1.54e-28 | 111 | 443 | 69 | 396 | ChainA, Glycoside hydrolase family 2, candidate beta-glycosidase [Phocaeicola vulgatus ATCC 8482] |
| 4YPJ_A | 3.30e-27 | 111 | 468 | 73 | 437 | ChainA, Beta galactosidase [Niallia circulans],4YPJ_B Chain B, Beta galactosidase [Niallia circulans] |
| 3FN9_A | 1.20e-26 | 67 | 443 | 14 | 400 | Crystalstructure of putative beta-galactosidase from bacteroides fragilis [Bacteroides fragilis NCTC 9343],3FN9_B Crystal structure of putative beta-galactosidase from bacteroides fragilis [Bacteroides fragilis NCTC 9343],3FN9_C Crystal structure of putative beta-galactosidase from bacteroides fragilis [Bacteroides fragilis NCTC 9343],3FN9_D Crystal structure of putative beta-galactosidase from bacteroides fragilis [Bacteroides fragilis NCTC 9343] |
| 6D50_A | 2.62e-26 | 53 | 443 | 28 | 426 | Bacteroidesuniforms beta-glucuronidase 2 bound to D-glucaro-1,5-lactone [Bacteroides uniformis str. 3978 T3 ii],6D50_B Bacteroides uniforms beta-glucuronidase 2 bound to D-glucaro-1,5-lactone [Bacteroides uniformis str. 3978 T3 ii] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P77989 | 6.14e-30 | 111 | 440 | 59 | 384 | Beta-galactosidase OS=Thermoanaerobacter pseudethanolicus (strain ATCC 33223 / 39E) OX=340099 GN=lacZ PE=3 SV=2 |
| T2KPJ7 | 3.92e-28 | 111 | 452 | 106 | 439 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_21970 PE=2 SV=1 |
| T2KM09 | 1.32e-25 | 111 | 468 | 109 | 452 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22050 PE=2 SV=2 |
| P23989 | 8.82e-25 | 24 | 559 | 40 | 611 | Beta-galactosidase OS=Streptococcus thermophilus OX=1308 GN=lacZ PE=3 SV=1 |
| Q02603 | 2.32e-23 | 112 | 453 | 132 | 476 | Beta-galactosidase large subunit OS=Leuconostoc lactis OX=1246 GN=lacL PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
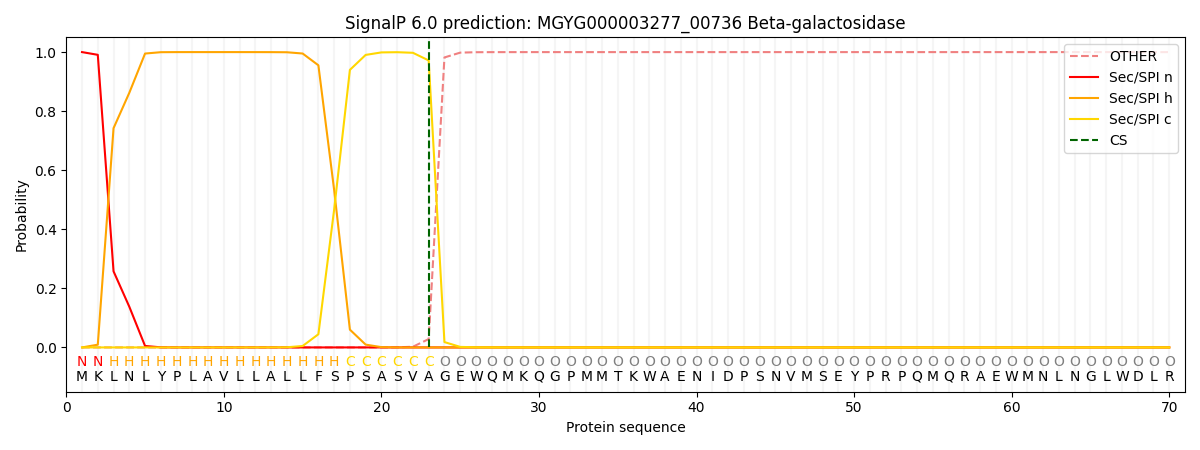
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000338 | 0.998939 | 0.000198 | 0.000182 | 0.000170 | 0.000165 |
