You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003282_02290
You are here: Home > Sequence: MGYG000003282_02290
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
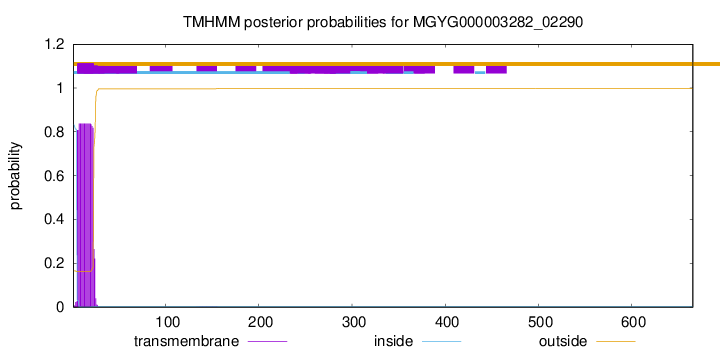
TMHMM annotations
Basic Information help
| Species | Coprobacter sp900545915 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Coprobacteraceae; Coprobacter; Coprobacter sp900545915 | |||||||||||
| CAZyme ID | MGYG000003282_02290 | |||||||||||
| CAZy Family | PL12 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 7439; End: 9439 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL12 | 420 | 541 | 9.5e-19 | 0.9420289855072463 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07940 | Hepar_II_III | 2.55e-18 | 412 | 556 | 5 | 159 | Heparinase II/III-like protein. This family features sequences that are similar to a region of the Flavobacterium heparinum proteins heparinase II and heparinase III. The former is known to degrade heparin and heparin sulphate, whereas the latter predominantly degrades heparin sulphate. Both are secreted into the periplasmic space upon induction with heparin. |
| pfam16889 | Hepar_II_III_N | 6.02e-08 | 283 | 352 | 243 | 317 | Heparinase II/III N-terminus. This is the N-terminal domain of heparinase II/III proteins. It is a toroid-like domain. |
| pfam05426 | Alginate_lyase | 3.01e-07 | 153 | 328 | 68 | 250 | Alginate lyase. This family contains several bacterial alginate lyase proteins. Alginate is a family of 1-4-linked copolymers of beta -D-mannuronic acid (M) and alpha -L-guluronic acid (G). It is produced by brown algae and by some bacteria belonging to the genera Azotobacter and Pseudomonas. Alginate lyases catalyze the depolymerization of alginates by beta -elimination, generating a molecule containing 4-deoxy-L-erythro-hex-4-enepyranosyluronate at the nonreducing end. This family adopts an all alpha fold. |
| cd00244 | AlgLyase | 0.002 | 223 | 327 | 148 | 259 | Alginate Lyase A1-III; enzymatically depolymerizes alginate, a complex copolymer of beta-D-mannuronate and alpha-L-guluronate, by cleaving the beta-(1,4) glycosidic bond. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| SCD20870.1 | 1.15e-155 | 42 | 659 | 20 | 633 |
| QDO68586.1 | 2.20e-140 | 40 | 659 | 17 | 633 |
| QUT88595.1 | 7.66e-138 | 40 | 659 | 17 | 633 |
| ALJ60398.1 | 7.66e-138 | 40 | 659 | 17 | 633 |
| QDO69002.1 | 6.38e-125 | 45 | 659 | 598 | 1213 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4NEI_A | 5.70e-24 | 139 | 559 | 88 | 549 | Alg17cPL17 Family Alginate Lyase [Saccharophagus degradans 2-40],4NEI_B Alg17c PL17 Family Alginate Lyase [Saccharophagus degradans 2-40] |
| 4OK4_A | 5.70e-24 | 139 | 559 | 88 | 549 | CrystalStructure of Alg17c Mutant H202L [Saccharophagus degradans 2-40],4OK4_B Crystal Structure of Alg17c Mutant H202L [Saccharophagus degradans 2-40] |
| 4OJZ_A | 1.00e-23 | 139 | 559 | 88 | 549 | CrystalStructure of Alg17c Mutant Y258A Complexed with Alginate Trisaccharide [Saccharophagus degradans 2-40],4OJZ_B Crystal Structure of Alg17c Mutant Y258A Complexed with Alginate Trisaccharide [Saccharophagus degradans 2-40],4OK2_A Crystal Structure of Alg17c Mutant Y258A [Saccharophagus degradans 2-40],4OK2_B Crystal Structure of Alg17c Mutant Y258A [Saccharophagus degradans 2-40] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B2FSW8 | 4.85e-26 | 139 | 559 | 95 | 554 | Alginate lyase OS=Stenotrophomonas maltophilia (strain K279a) OX=522373 GN=Smlt2602 PE=1 SV=1 |
| Q21FJ0 | 3.12e-23 | 139 | 559 | 88 | 549 | Exo-oligoalginate lyase OS=Saccharophagus degradans (strain 2-40 / ATCC 43961 / DSM 17024) OX=203122 GN=alg17C PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
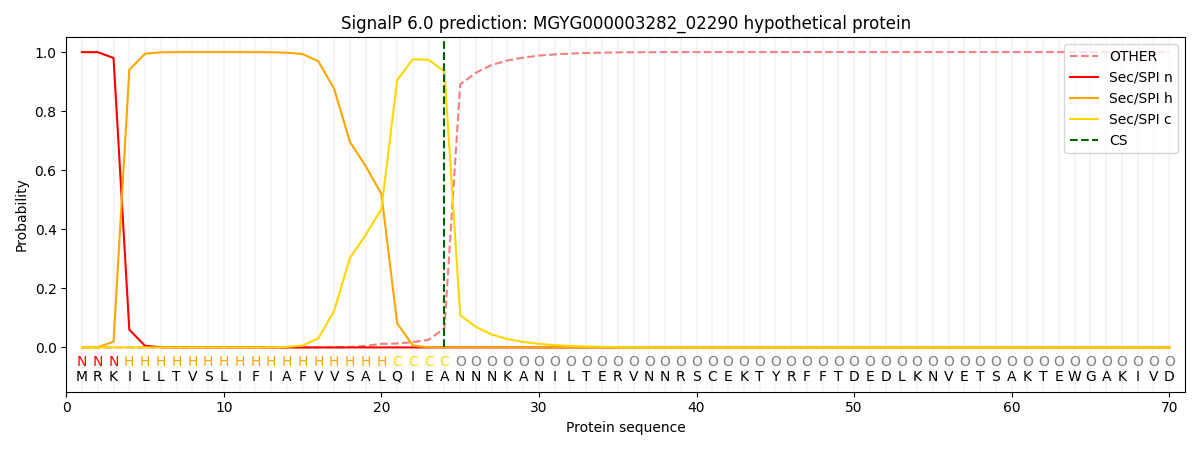
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000444 | 0.998882 | 0.000202 | 0.000160 | 0.000152 | 0.000147 |