You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003312_03752
You are here: Home > Sequence: MGYG000003312_03752
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides sp900547205 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides sp900547205 | |||||||||||
| CAZyme ID | MGYG000003312_03752 | |||||||||||
| CAZy Family | GH76 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 87743; End: 88945 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH76 | 66 | 382 | 5.5e-70 | 0.8547486033519553 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam03663 | Glyco_hydro_76 | 7.41e-30 | 86 | 304 | 34 | 256 | Glycosyl hydrolase family 76. Family of alpha-1,6-mannanases. |
| COG4833 | COG4833 | 2.33e-11 | 144 | 400 | 88 | 352 | Predicted alpha-1,6-mannanase, GH76 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGT72765.1 | 5.20e-246 | 1 | 399 | 1 | 399 |
| QDO71507.1 | 1.95e-234 | 1 | 399 | 1 | 399 |
| ALJ59939.1 | 1.13e-233 | 1 | 399 | 1 | 399 |
| QUT89047.1 | 4.58e-233 | 1 | 399 | 1 | 399 |
| BAD50241.1 | 2.09e-215 | 1 | 399 | 1 | 399 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4MU9_A | 4.44e-102 | 37 | 393 | 8 | 360 | Crystalstructure of a putative glycosylhydrolase (BT_3782) from Bacteroides thetaiotaomicron VPI-5482 at 1.89 A resolution [Bacteroides thetaiotaomicron VPI-5482],4MU9_B Crystal structure of a putative glycosylhydrolase (BT_3782) from Bacteroides thetaiotaomicron VPI-5482 at 1.89 A resolution [Bacteroides thetaiotaomicron VPI-5482] |
| 4C1S_A | 5.13e-67 | 87 | 399 | 45 | 371 | Glycosidehydrolase family 76 (mannosidase) Bt3792 from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482],4C1S_B Glycoside hydrolase family 76 (mannosidase) Bt3792 from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482] |
| 6U4Z_A | 1.50e-63 | 53 | 396 | 137 | 492 | CrystalStructure of a family 76 glycoside hydrolase from a bovine Bacteroides thetaiotaomicron strain [Bacteroides thetaiotaomicron] |
| 4V1S_A | 1.56e-30 | 125 | 367 | 113 | 357 | Structureof the GH76 alpha-mannanase BT2949 from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482],4V1S_B Structure of the GH76 alpha-mannanase BT2949 from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482] |
| 4V1R_A | 3.52e-28 | 125 | 367 | 113 | 357 | Structureof a selenomethionine derivative of the GH76 alpha- mannanase BT2949 Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482],4V1R_B Structure of a selenomethionine derivative of the GH76 alpha- mannanase BT2949 Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q5ACZ2 | 5.24e-06 | 166 | 312 | 140 | 299 | Mannan endo-1,6-alpha-mannosidase DFG5 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=DFG5 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
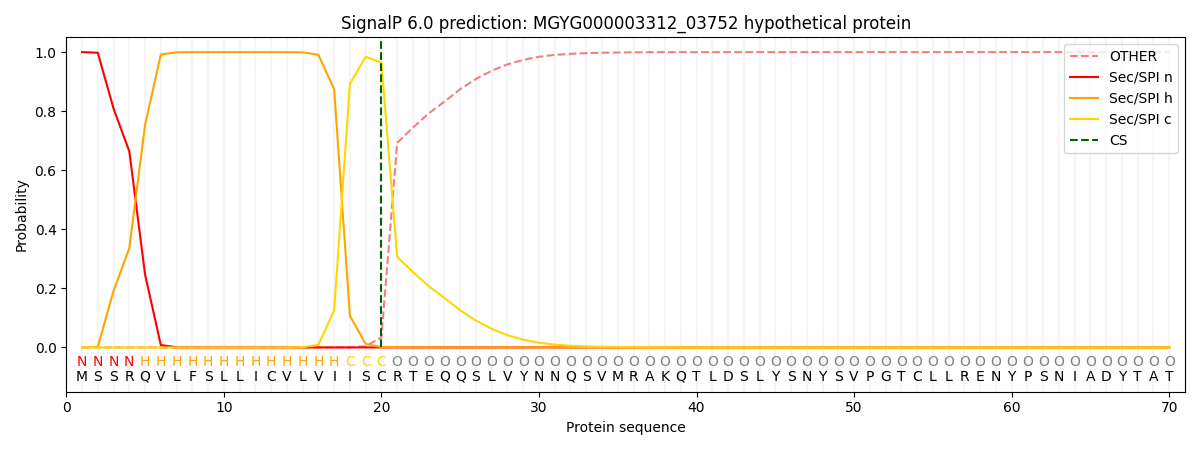
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000339 | 0.998938 | 0.000285 | 0.000139 | 0.000135 | 0.000129 |
