You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003335_00359
Basic Information
help
| Species |
GCA-900066135 sp900543575
|
| Lineage |
Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; GCA-900066135; GCA-900066135 sp900543575
|
| CAZyme ID |
MGYG000003335_00359
|
| CAZy Family |
GH141 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000003335 |
3295822 |
MAG |
China |
Asia |
|
| Gene Location |
Start: 12672;
End: 14990
Strand: -
|
No EC number prediction in MGYG000003335_00359.
| Family |
Start |
End |
Evalue |
family coverage |
| GH141 |
8 |
462 |
2.3e-40 |
0.8633776091081594 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| pfam13229
|
Beta_helix |
8.56e-06 |
276 |
453 |
1 |
136 |
Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam13229
|
Beta_helix |
0.002 |
393 |
553 |
13 |
136 |
Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam13229
|
Beta_helix |
0.003 |
271 |
329 |
19 |
76 |
Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
This protein is predicted as OTHER
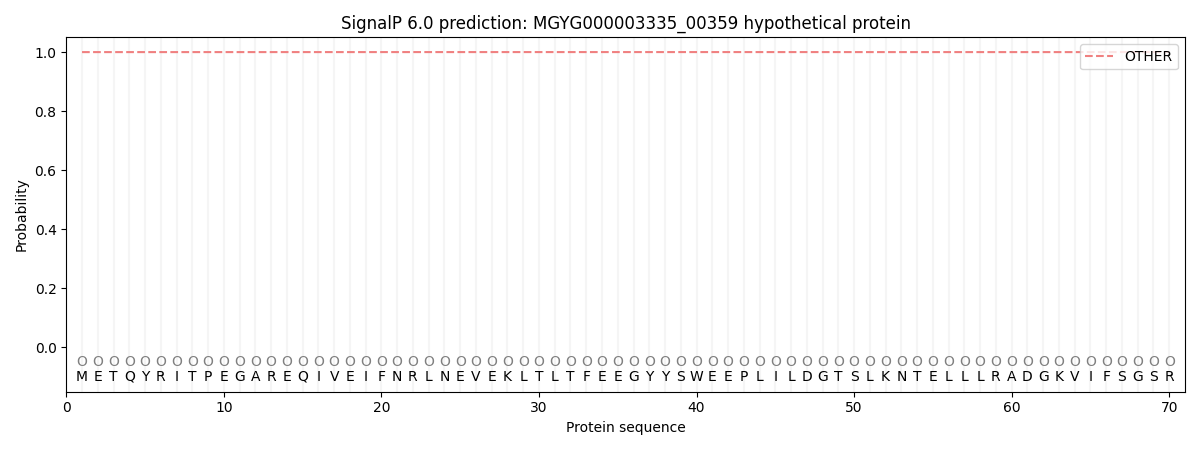
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
1.000062
|
0.000000
|
0.000000
|
0.000000
|
0.000000
|
0.000000
|
There is no transmembrane helices in MGYG000003335_00359.

