You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003337_02111
You are here: Home > Sequence: MGYG000003337_02111
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Ruminococcus_C sp900765125 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Ruminococcus_C; Ruminococcus_C sp900765125 | |||||||||||
| CAZyme ID | MGYG000003337_02111 | |||||||||||
| CAZy Family | GH9 | |||||||||||
| CAZyme Description | Endoglucanase E-4 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 14234; End: 16714 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH9 | 39 | 488 | 1.4e-104 | 0.9952153110047847 |
| CBM3 | 518 | 607 | 5.4e-16 | 0.9318181818181818 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00759 | Glyco_hydro_9 | 2.97e-98 | 42 | 488 | 1 | 374 | Glycosyl hydrolase family 9. |
| PLN02420 | PLN02420 | 7.28e-61 | 13 | 481 | 14 | 495 | endoglucanase |
| PLN02340 | PLN02340 | 7.46e-59 | 36 | 509 | 27 | 508 | endoglucanase |
| PLN02345 | PLN02345 | 5.20e-57 | 43 | 490 | 1 | 456 | endoglucanase |
| PLN02613 | PLN02613 | 1.34e-55 | 39 | 492 | 26 | 479 | endoglucanase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BAJ05816.1 | 0.0 | 5 | 692 | 4 | 696 |
| ADU22928.1 | 0.0 | 5 | 692 | 4 | 696 |
| CAL91976.1 | 9.90e-278 | 39 | 694 | 11 | 634 |
| CAL91977.1 | 5.12e-269 | 39 | 694 | 30 | 639 |
| CBL17683.1 | 2.08e-246 | 38 | 809 | 41 | 727 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5GXX_A | 7.08e-155 | 38 | 690 | 5 | 602 | ChainA, Glucanase [Acetivibrio thermocellus],5GXX_B Chain B, Glucanase [Acetivibrio thermocellus],5GXY_A Chain A, Glucanase [Acetivibrio thermocellus],5GXY_B Chain B, Glucanase [Acetivibrio thermocellus],5GXZ_A Chain A, Glucanase [Acetivibrio thermocellus],5GXZ_B Chain B, Glucanase [Acetivibrio thermocellus] |
| 5GY0_A | 5.57e-154 | 38 | 690 | 5 | 602 | ChainA, Glucanase [Acetivibrio thermocellus],5GY0_B Chain B, Glucanase [Acetivibrio thermocellus] |
| 5GY1_A | 1.11e-153 | 38 | 690 | 5 | 602 | ChainA, Glucanase [Acetivibrio thermocellus],5GY1_B Chain B, Glucanase [Acetivibrio thermocellus] |
| 1G87_A | 3.43e-143 | 38 | 688 | 4 | 614 | TheCrystal Structure Of Endoglucanase 9g From Clostridium Cellulolyticum [Ruminiclostridium cellulolyticum],1G87_B The Crystal Structure Of Endoglucanase 9g From Clostridium Cellulolyticum [Ruminiclostridium cellulolyticum],1GA2_A The Crystal Structure Of Endoglucanase 9g From Clostridium Cellulolyticum Complexed With Cellobiose [Ruminiclostridium cellulolyticum],1GA2_B The Crystal Structure Of Endoglucanase 9g From Clostridium Cellulolyticum Complexed With Cellobiose [Ruminiclostridium cellulolyticum],1KFG_A The X-ray Crystal Structure of Cel9G from Clostridium cellulolyticum complexed with a Thio-Oligosaccharide Inhibitor [Ruminiclostridium cellulolyticum],1KFG_B The X-ray Crystal Structure of Cel9G from Clostridium cellulolyticum complexed with a Thio-Oligosaccharide Inhibitor [Ruminiclostridium cellulolyticum] |
| 1K72_A | 4.84e-143 | 38 | 688 | 4 | 614 | TheX-ray Crystal Structure Of Cel9G Complexed With cellotriose [Ruminiclostridium cellulolyticum],1K72_B The X-ray Crystal Structure Of Cel9G Complexed With cellotriose [Ruminiclostridium cellulolyticum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P37700 | 2.38e-142 | 1 | 690 | 2 | 651 | Endoglucanase G OS=Ruminiclostridium cellulolyticum (strain ATCC 35319 / DSM 5812 / JCM 6584 / H10) OX=394503 GN=celCCG PE=1 SV=2 |
| P26221 | 1.47e-137 | 32 | 689 | 44 | 652 | Endoglucanase E-4 OS=Thermobifida fusca OX=2021 GN=celD PE=1 SV=2 |
| P22534 | 1.68e-133 | 38 | 688 | 26 | 636 | Endoglucanase A OS=Caldicellulosiruptor saccharolyticus OX=44001 GN=celA PE=3 SV=2 |
| P26224 | 8.14e-132 | 3 | 789 | 1 | 699 | Endoglucanase F OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celF PE=3 SV=1 |
| Q02934 | 7.22e-131 | 38 | 688 | 76 | 683 | Endoglucanase 1 OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celI PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
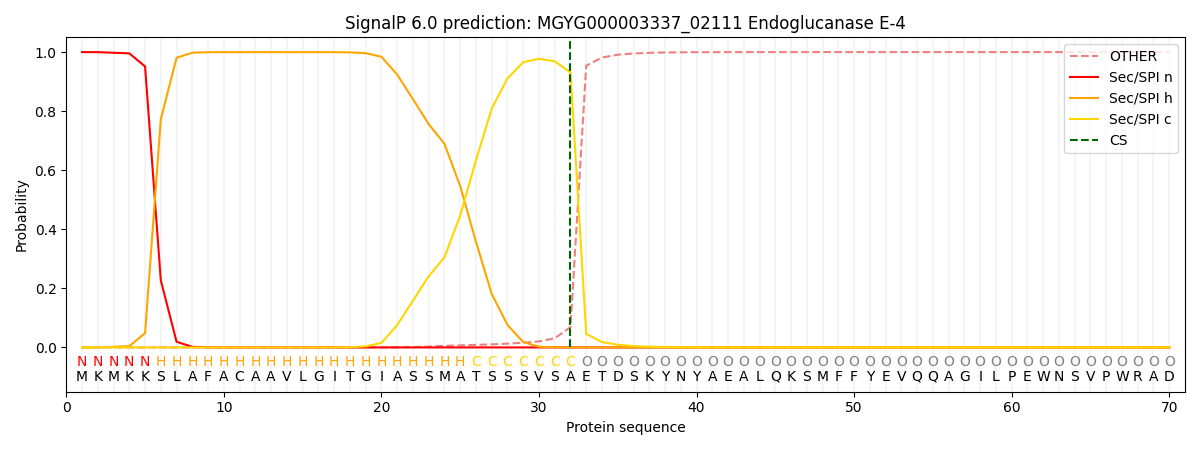
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000375 | 0.998611 | 0.000427 | 0.000213 | 0.000178 | 0.000156 |
