You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003351_03777
You are here: Home > Sequence: MGYG000003351_03777
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides sp900765785 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides sp900765785 | |||||||||||
| CAZyme ID | MGYG000003351_03777 | |||||||||||
| CAZy Family | PL9 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 36517; End: 38112 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL9 | 20 | 436 | 6.6e-106 | 0.9652406417112299 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam16380 | DUF4990 | 1.40e-58 | 382 | 522 | 1 | 141 | Domain of unknown function. This family around 150 residues locates in the C-terminal of some uncharacterized proteins in various Bacteroides species. The function of this family remains unknown. |
| pfam13229 | Beta_helix | 1.57e-06 | 161 | 332 | 4 | 157 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam07602 | DUF1565 | 7.42e-04 | 32 | 67 | 3 | 38 | Protein of unknown function (DUF1565). These proteins share a region of homology in their N termini, and are found in several phylogenetically diverse bacteria and in the archaeon Methanosarcina acetivorans. Some of these proteins also contain characterized domains such as pfam00395 and pfam03422. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT42065.1 | 0.0 | 1 | 531 | 1 | 531 |
| QUT69867.1 | 0.0 | 1 | 531 | 1 | 531 |
| ALJ43466.1 | 3.45e-286 | 1 | 531 | 1 | 535 |
| QMW86514.1 | 3.45e-286 | 1 | 531 | 1 | 535 |
| AAO79275.1 | 3.45e-286 | 1 | 531 | 1 | 535 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5OLQ_A | 1.90e-274 | 24 | 531 | 3 | 514 | Rhamnogalacturonanlyase [Bacteroides thetaiotaomicron],5OLQ_B Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLQ_C Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLR_A Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLR_B Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLR_C Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLS_A Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron] |
| 1RU4_A | 1.46e-45 | 16 | 436 | 7 | 375 | ChainA, Pectate lyase [Dickeya chrysanthemi] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P0C1A6 | 7.07e-45 | 16 | 436 | 32 | 400 | Pectate lyase L OS=Dickeya chrysanthemi OX=556 GN=pelL PE=3 SV=1 |
| P0C1A7 | 1.36e-44 | 16 | 436 | 32 | 400 | Pectate lyase L OS=Dickeya dadantii (strain 3937) OX=198628 GN=pelL PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
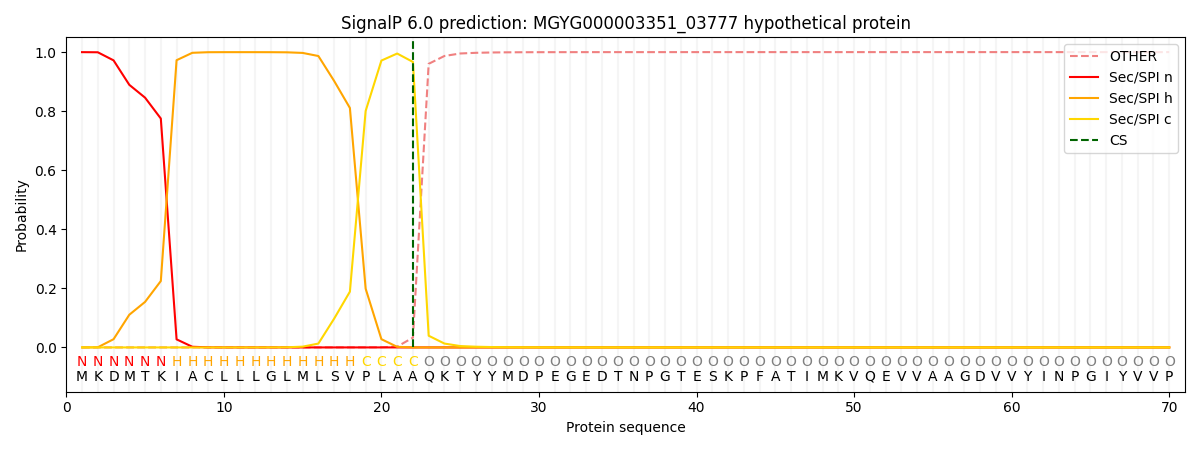
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000475 | 0.998311 | 0.000586 | 0.000208 | 0.000197 | 0.000187 |
