You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003355_01965
You are here: Home > Sequence: MGYG000003355_01965
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Lachnoclostridium phytofermentans_A | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Lachnoclostridium; Lachnoclostridium phytofermentans_A | |||||||||||
| CAZyme ID | MGYG000003355_01965 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 31544; End: 35317 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4193 | LytD | 3.73e-21 | 910 | 1147 | 45 | 244 | Beta- N-acetylglucosaminidase [Carbohydrate transport and metabolism]. |
| pfam08239 | SH3_3 | 1.33e-08 | 330 | 389 | 1 | 54 | Bacterial SH3 domain. |
| pfam08239 | SH3_3 | 5.14e-08 | 142 | 201 | 1 | 54 | Bacterial SH3 domain. |
| pfam08239 | SH3_3 | 9.36e-07 | 526 | 585 | 1 | 54 | Bacterial SH3 domain. |
| NF033186 | internalin_K | 5.13e-05 | 402 | 522 | 438 | 559 | class 1 internalin InlK. Internalins, as found in the intracellular human pathogen Listeria monocytogenes, are paralogous surface-anchored proteins with an N-terminal signal peptide, leucine-rich repeats, and a C-terminal LPXTG processing and cell surface anchoring site. Members of this family are internalin K (InlK), a virulence factor. See articles PMID:17764999. for a general discussion of internalins, and PMID:21829365, PMID:22082958, and PMID:23958637 for more information about internalin K. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ABX43817.1 | 0.0 | 1 | 1257 | 15 | 1281 |
| BCJ93456.1 | 2.74e-218 | 421 | 1255 | 60 | 895 |
| QHQ61172.1 | 4.24e-200 | 516 | 1255 | 134 | 767 |
| CUH91961.1 | 6.34e-199 | 523 | 1255 | 144 | 912 |
| BCJ97942.1 | 2.48e-198 | 429 | 1255 | 60 | 907 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6FXO_A | 4.01e-12 | 983 | 1147 | 92 | 243 | ChainA, Bifunctional autolysin [Staphylococcus aureus subsp. aureus Mu50] |
| 4PI7_A | 6.03e-08 | 983 | 1136 | 84 | 216 | ChainA, Autolysin E [Staphylococcus aureus subsp. aureus Mu50],4PI9_A Chain A, Autolysin E [Staphylococcus aureus subsp. aureus Mu50],4PIA_A Chain A, Autolysin E [Staphylococcus aureus subsp. aureus Mu50] |
| 4PI8_A | 3.58e-07 | 983 | 1136 | 84 | 216 | ChainA, Autolysin E [Staphylococcus aureus subsp. aureus Mu50] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8CPQ1 | 2.11e-12 | 913 | 1147 | 1128 | 1334 | Bifunctional autolysin OS=Staphylococcus epidermidis (strain ATCC 12228 / FDA PCI 1200) OX=176280 GN=atl PE=3 SV=1 |
| O33635 | 4.75e-12 | 913 | 1147 | 1128 | 1334 | Bifunctional autolysin OS=Staphylococcus epidermidis OX=1282 GN=atl PE=1 SV=1 |
| Q5HQB9 | 4.75e-12 | 913 | 1147 | 1128 | 1334 | Bifunctional autolysin OS=Staphylococcus epidermidis (strain ATCC 35984 / RP62A) OX=176279 GN=atl PE=3 SV=1 |
| Q99V41 | 2.71e-10 | 983 | 1147 | 1096 | 1247 | Bifunctional autolysin OS=Staphylococcus aureus (strain N315) OX=158879 GN=atl PE=1 SV=1 |
| Q6GI31 | 2.71e-10 | 983 | 1147 | 1105 | 1256 | Bifunctional autolysin OS=Staphylococcus aureus (strain MRSA252) OX=282458 GN=atl PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
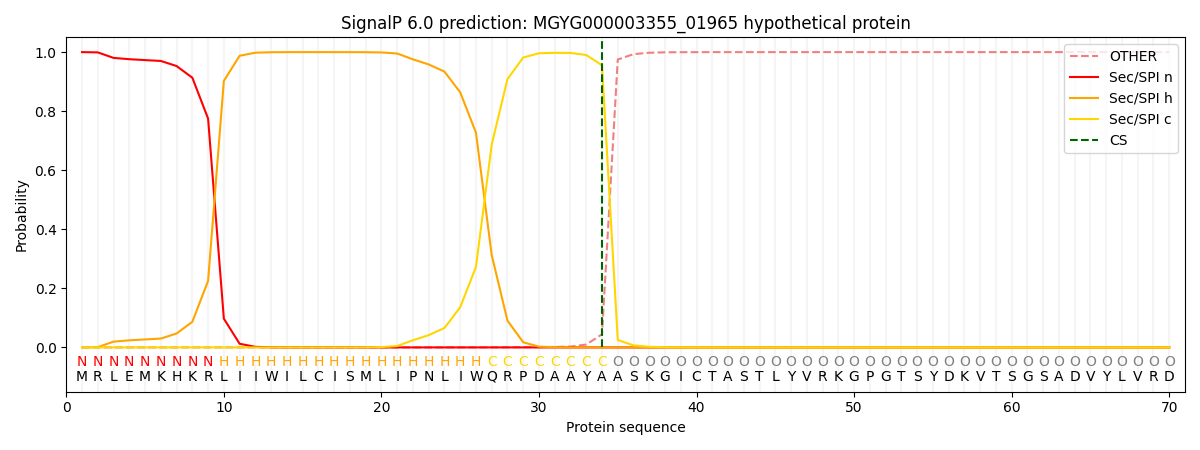
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000379 | 0.998901 | 0.000191 | 0.000193 | 0.000152 | 0.000145 |
