You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003355_03656
You are here: Home > Sequence: MGYG000003355_03656
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
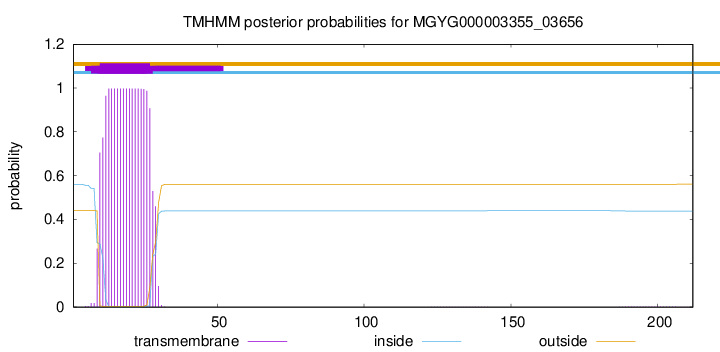
TMHMM annotations
Basic Information help
| Species | Lachnoclostridium phytofermentans_A | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Lachnoclostridium; Lachnoclostridium phytofermentans_A | |||||||||||
| CAZyme ID | MGYG000003355_03656 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 14018; End: 14656 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM32 | 81 | 209 | 4.5e-17 | 0.9435483870967742 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00754 | F5_F8_type_C | 7.71e-13 | 83 | 208 | 4 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| cd00057 | FA58C | 2.53e-09 | 74 | 211 | 6 | 142 | Substituted updates: Jan 31, 2002 |
| smart00231 | FA58C | 1.38e-04 | 75 | 211 | 3 | 137 | Coagulation factor 5/8 C-terminal domain, discoidin domain. Cell surface-attached carbohydrate-binding domain, present in eukaryotes and assumed to have horizontally transferred to eubacterial genomes. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ABX43663.1 | 4.80e-129 | 5 | 212 | 1 | 208 |
| BCK00430.1 | 1.16e-68 | 10 | 211 | 7 | 205 |
| ACK41503.1 | 3.16e-37 | 31 | 211 | 495 | 679 |
| ACI19833.1 | 2.64e-35 | 44 | 211 | 512 | 679 |
| AEJ61831.1 | 8.31e-32 | 75 | 211 | 25 | 159 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER
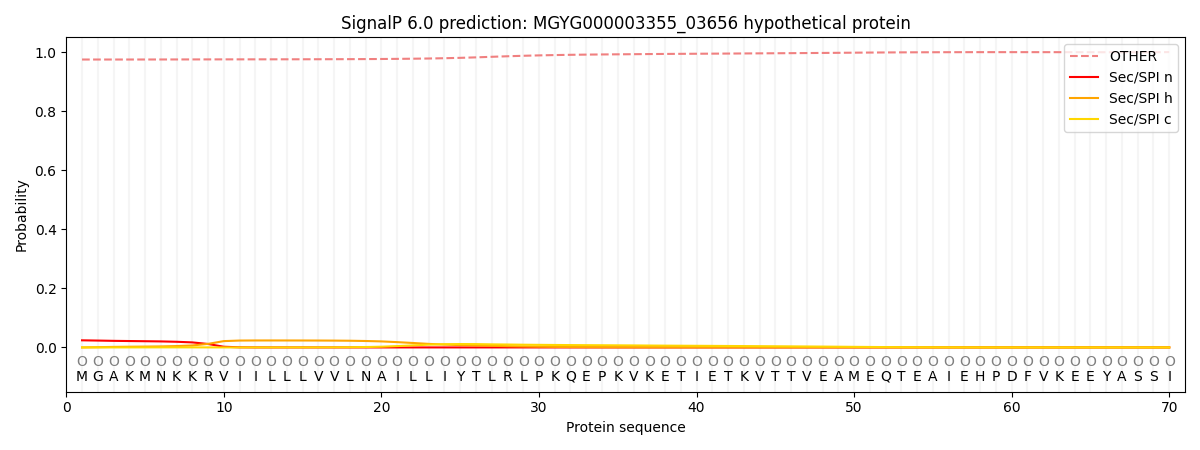
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.976235 | 0.022248 | 0.001107 | 0.000104 | 0.000059 | 0.000272 |