You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003355_03657
You are here: Home > Sequence: MGYG000003355_03657
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Lachnoclostridium phytofermentans_A | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Lachnoclostridium; Lachnoclostridium phytofermentans_A | |||||||||||
| CAZyme ID | MGYG000003355_03657 | |||||||||||
| CAZy Family | GH28 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 14808; End: 16364 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH28 | 225 | 496 | 5.9e-42 | 0.7630769230769231 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5434 | Pgu1 | 8.62e-11 | 221 | 351 | 178 | 336 | Polygalacturonase [Carbohydrate transport and metabolism]. |
| pfam00295 | Glyco_hydro_28 | 5.15e-09 | 237 | 419 | 50 | 238 | Glycosyl hydrolases family 28. Glycosyl hydrolase family 28 includes polygalacturonase EC:3.2.1.15 as well as rhamnogalacturonase A(RGase A), EC:3.2.1.-. These enzymes are important in cell wall metabolism. |
| PLN02793 | PLN02793 | 3.97e-08 | 229 | 419 | 140 | 329 | Probable polygalacturonase |
| PLN02218 | PLN02218 | 1.09e-06 | 212 | 498 | 150 | 414 | polygalacturonase ADPG |
| PLN02188 | PLN02188 | 3.69e-04 | 238 | 415 | 122 | 344 | polygalacturonase/glycoside hydrolase family protein |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ABX43664.1 | 0.0 | 1 | 518 | 1 | 518 |
| BCK00431.1 | 1.36e-238 | 7 | 518 | 3 | 520 |
| ACK41503.1 | 1.87e-145 | 39 | 508 | 27 | 475 |
| ACI19833.1 | 1.05e-144 | 39 | 508 | 27 | 475 |
| QTE67472.1 | 1.42e-134 | 45 | 508 | 33 | 478 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3JUR_A | 4.54e-06 | 231 | 342 | 116 | 270 | Thecrystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_B The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_C The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_D The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER
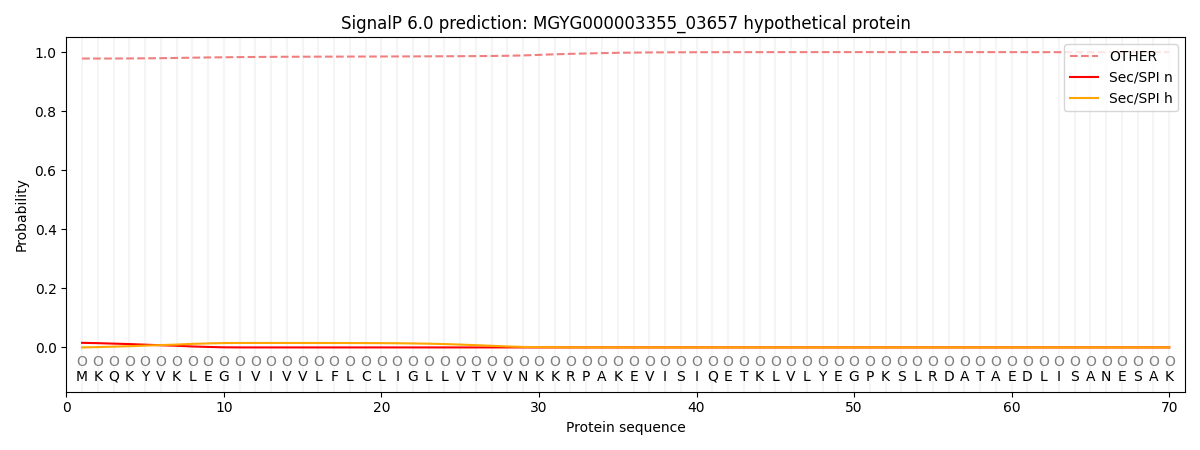
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.979020 | 0.014344 | 0.000367 | 0.000083 | 0.000054 | 0.006160 |

