You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003359_00457
You are here: Home > Sequence: MGYG000003359_00457
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
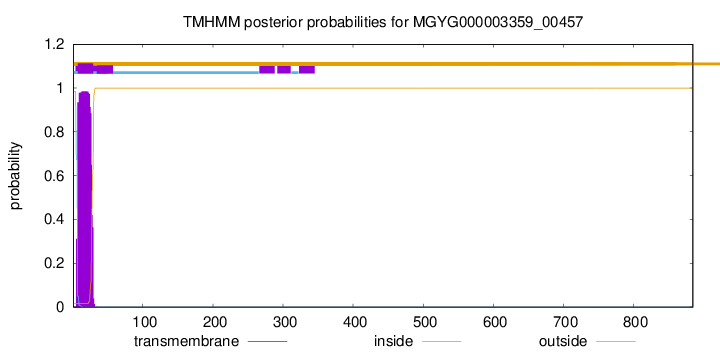
TMHMM annotations
Basic Information help
| Species | Paenibacillus_A sp900766135 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus_A; Paenibacillus_A sp900766135 | |||||||||||
| CAZyme ID | MGYG000003359_00457 | |||||||||||
| CAZy Family | PL29 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 300855; End: 303512 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL29 | 92 | 378 | 4.6e-25 | 0.9169435215946844 |
| CBM32 | 767 | 880 | 3.3e-17 | 0.9032258064516129 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00754 | F5_F8_type_C | 2.06e-13 | 764 | 880 | 1 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam12708 | Pectate_lyase_3 | 9.32e-11 | 92 | 160 | 2 | 62 | Pectate lyase superfamily protein. This family of proteins possesses a beta helical structure like Pectate lyase. This family is most closely related to glycosyl hydrolase family 28. |
| COG5434 | Pgu1 | 4.06e-06 | 80 | 135 | 66 | 124 | Polygalacturonase [Carbohydrate transport and metabolism]. |
| cd00057 | FA58C | 5.61e-05 | 782 | 880 | 32 | 137 | Substituted updates: Jan 31, 2002 |
| smart00231 | FA58C | 1.20e-04 | 777 | 872 | 26 | 129 | Coagulation factor 5/8 C-terminal domain, discoidin domain. Cell surface-attached carbohydrate-binding domain, present in eukaryotes and assumed to have horizontally transferred to eubacterial genomes. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QLG40566.1 | 0.0 | 1 | 884 | 1 | 887 |
| QOS79355.1 | 0.0 | 1 | 883 | 1 | 886 |
| QAY65253.1 | 0.0 | 42 | 883 | 41 | 885 |
| QNK56161.1 | 1.49e-212 | 43 | 883 | 261 | 1110 |
| QRO01698.1 | 3.98e-128 | 48 | 566 | 31 | 546 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7D29_A | 1.74e-25 | 762 | 884 | 11 | 134 | CBM32of AlyQ [Persicobacter sp. CCB-QB2],7D2A_A CBM32 of AlyQ in complex with 4,5-unsaturated mannuronic acid [Persicobacter sp. CCB-QB2] |
| 5XNR_A | 3.20e-23 | 762 | 884 | 11 | 134 | TruncatedAlyQ with CBM32 and alginate lyase domains [Persicobacter sp. CCB-QB2] |
| 5ZU6_A | 6.31e-21 | 763 | 877 | 35 | 151 | ACBM32 derived from alginate lyase B (AlyB-OU02) [Vibrio] |
| 5ZU5_A | 8.92e-19 | 763 | 877 | 35 | 151 | Crystalstructure of a full length alginate lyase with CBM domain [Vibrio splendidus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9ZFG9 | 2.69e-06 | 99 | 185 | 8 | 95 | Alginate lyase 7 OS=Azotobacter vinelandii OX=354 GN=algE7 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000505 | 0.998581 | 0.000364 | 0.000192 | 0.000169 | 0.000148 |