You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003359_03066
You are here: Home > Sequence: MGYG000003359_03066
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Paenibacillus_A sp900766135 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus_A; Paenibacillus_A sp900766135 | |||||||||||
| CAZyme ID | MGYG000003359_03066 | |||||||||||
| CAZy Family | GH141 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 84844; End: 88650 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH141 | 48 | 336 | 6.6e-35 | 0.4629981024667932 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02368 | Big_2 | 3.01e-11 | 751 | 826 | 1 | 77 | Bacterial Ig-like domain (group 2). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in bacterial and phage surface proteins such as intimins. |
| COG5492 | YjdB | 1.65e-10 | 749 | 858 | 180 | 289 | Uncharacterized conserved protein YjdB, contains Ig-like domain [General function prediction only]. |
| pfam13229 | Beta_helix | 6.55e-10 | 397 | 590 | 1 | 156 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam13229 | Beta_helix | 3.07e-09 | 375 | 567 | 1 | 156 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam13229 | Beta_helix | 3.62e-09 | 353 | 506 | 3 | 131 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNK56945.1 | 5.71e-116 | 49 | 735 | 41 | 731 |
| QUH28384.1 | 4.88e-109 | 48 | 735 | 40 | 726 |
| QUT84604.1 | 5.50e-63 | 167 | 734 | 160 | 719 |
| AUP78440.1 | 6.72e-61 | 45 | 748 | 24 | 695 |
| QNN23113.1 | 2.03e-47 | 49 | 325 | 39 | 335 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P94576 | 2.75e-08 | 49 | 133 | 37 | 107 | Uncharacterized protein YwoF OS=Bacillus subtilis (strain 168) OX=224308 GN=ywoF PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
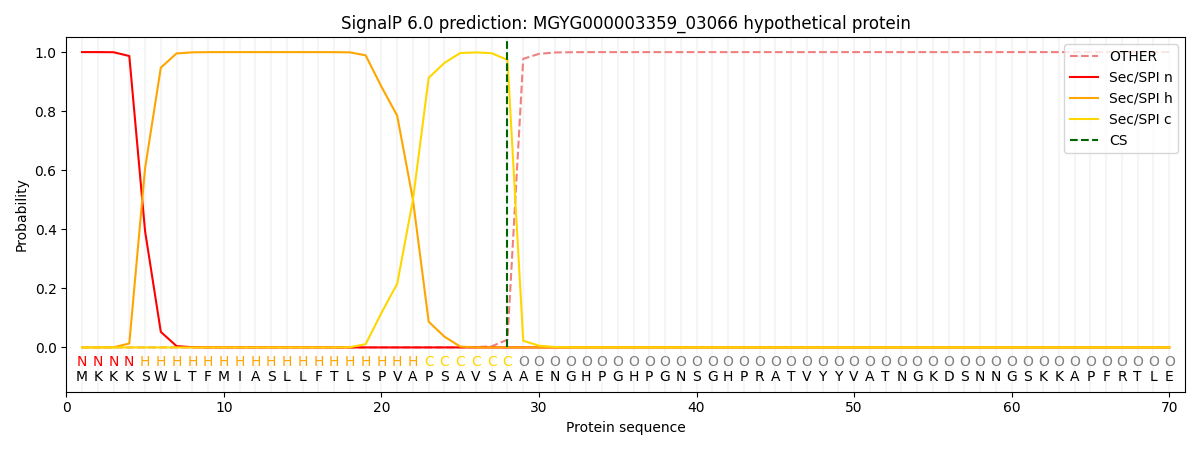
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000271 | 0.998967 | 0.000180 | 0.000206 | 0.000186 | 0.000161 |
