You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003362_02411
You are here: Home > Sequence: MGYG000003362_02411
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
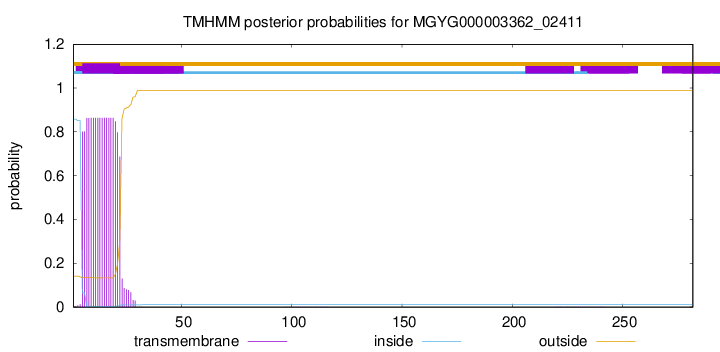
TMHMM annotations
Basic Information help
| Species | Dysgonomonas sp900079735 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Dysgonomonadaceae; Dysgonomonas; Dysgonomonas sp900079735 | |||||||||||
| CAZyme ID | MGYG000003362_02411 | |||||||||||
| CAZy Family | CE6 | |||||||||||
| CAZyme Description | Carbohydrate acetyl esterase/feruloyl esterase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 198711; End: 199559 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE6 | 87 | 199 | 7.2e-34 | 0.98989898989899 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam03629 | SASA | 6.36e-42 | 24 | 273 | 1 | 226 | Carbohydrate esterase, sialic acid-specific acetylesterase. The catalytic triad of this esterase enzyme comprises residues Ser127, His403 and Asp391 in UniProtKB:P70665. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QEK50423.1 | 6.52e-130 | 21 | 274 | 21 | 274 |
| QRX64991.1 | 5.50e-129 | 26 | 274 | 2 | 250 |
| ADQ79784.1 | 1.17e-126 | 1 | 279 | 1 | 279 |
| AMP99863.1 | 6.95e-126 | 1 | 274 | 14 | 288 |
| AOZ99799.1 | 5.17e-124 | 24 | 280 | 26 | 280 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| D5EXZ4 | 3.21e-78 | 24 | 269 | 45 | 289 | Carbohydrate acetyl esterase/feruloyl esterase OS=Prevotella ruminicola (strain ATCC 19189 / JCM 8958 / 23) OX=264731 GN=axe1-6A PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
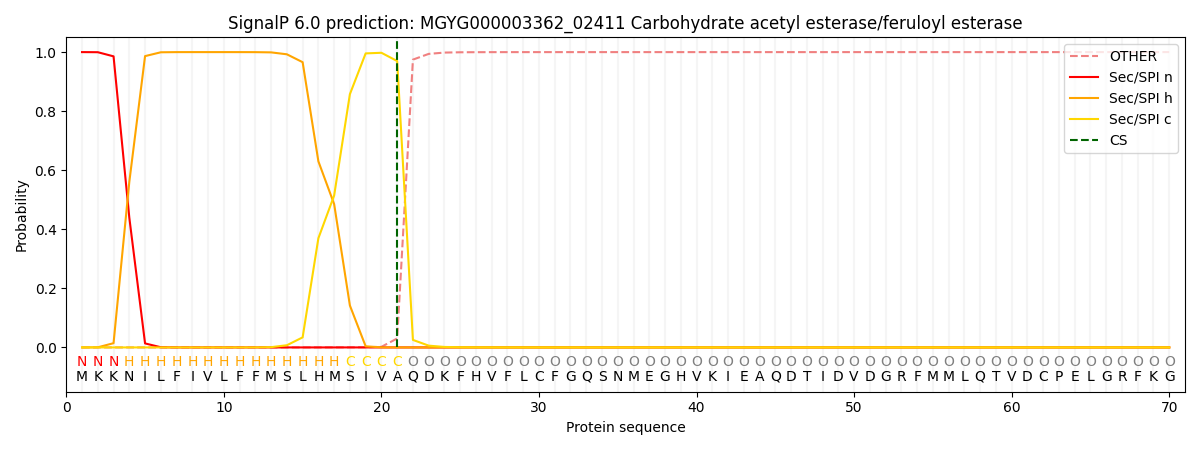
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000254 | 0.999134 | 0.000164 | 0.000144 | 0.000127 | 0.000130 |