You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003363_02200
You are here: Home > Sequence: MGYG000003363_02200
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
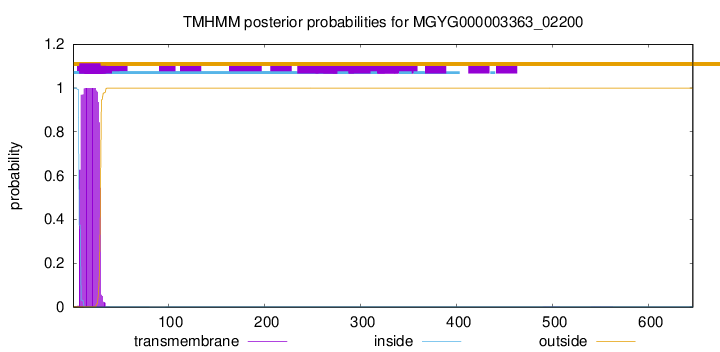
TMHMM annotations
Basic Information help
| Species | Bacteroides sp900766195 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides sp900766195 | |||||||||||
| CAZyme ID | MGYG000003363_02200 | |||||||||||
| CAZy Family | GT51 | |||||||||||
| CAZyme Description | Monofunctional biosynthetic peptidoglycan transglycosylase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 64791; End: 66731 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT51 | 440 | 590 | 2.9e-41 | 0.8361581920903954 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK00056 | mtgA | 1.41e-48 | 446 | 588 | 64 | 204 | monofunctional biosynthetic peptidoglycan transglycosylase; Provisional |
| pfam00912 | Transgly | 3.61e-44 | 428 | 613 | 1 | 177 | Transglycosylase. The penicillin-binding proteins are bifunctional proteins consisting of transglycosylase and transpeptidase in the N- and C-terminus respectively. The transglycosylase domain catalyzes the polymerization of murein glycan chains. |
| TIGR02070 | mono_pep_trsgly | 4.68e-42 | 429 | 587 | 45 | 198 | monofunctional biosynthetic peptidoglycan transglycosylase. This family is one of the transglycosylases involved in the late stages of peptidoglycan biosynthesis. Members tend to be small, about 240 amino acids in length, and consist almost entirely of a domain described by pfam00912 for transglycosylases. Species with this protein will have several other transglycosylases as well. All species with this protein are Proteobacteria that produce murein (peptidoglycan). [Cell envelope, Biosynthesis and degradation of murein sacculus and peptidoglycan] |
| COG0744 | MrcB | 4.86e-37 | 446 | 625 | 80 | 253 | Membrane carboxypeptidase (penicillin-binding protein) [Cell wall/membrane/envelope biogenesis]. |
| COG5009 | MrcA | 1.06e-24 | 444 | 639 | 69 | 260 | Membrane carboxypeptidase/penicillin-binding protein [Cell wall/membrane/envelope biogenesis]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QDO71568.1 | 5.01e-282 | 19 | 646 | 3 | 629 |
| ALJ61545.1 | 1.84e-272 | 19 | 646 | 3 | 629 |
| QUT92885.1 | 1.22e-270 | 19 | 646 | 3 | 629 |
| AAO77608.1 | 5.03e-270 | 2 | 636 | 3 | 630 |
| QMW84947.1 | 5.03e-270 | 2 | 636 | 3 | 630 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3NB6_A | 3.04e-20 | 437 | 624 | 15 | 195 | Crystalstructure of Aquifex aeolicus peptidoglycan glycosyltransferase in complex with Methylphosphoryl Neryl Moenomycin [Aquifex aeolicus] |
| 2OQO_A | 1.04e-19 | 437 | 624 | 15 | 195 | Crystalstructure of a peptidoglycan glycosyltransferase from a class A PBP: insight into bacterial cell wall synthesis [Aquifex aeolicus VF5],3D3H_A Crystal structure of a complex of the peptidoglycan glycosyltransferase domain from Aquifex aeolicus and neryl moenomycin A [Aquifex aeolicus],3NB7_A Crystal structure of Aquifex Aeolicus Peptidoglycan Glycosyltransferase in complex with Decarboxylated Neryl Moenomycin [Aquifex aeolicus] |
| 5U2G_A | 4.86e-14 | 459 | 645 | 57 | 238 | 2.6Angstrom Resolution Crystal Structure of Penicillin-Binding Protein 1A from Haemophilus influenzae [Haemophilus influenzae Rd KW20],5U2G_B 2.6 Angstrom Resolution Crystal Structure of Penicillin-Binding Protein 1A from Haemophilus influenzae [Haemophilus influenzae Rd KW20] |
| 7U4H_A | 4.19e-12 | 447 | 587 | 45 | 181 | ChainA, Penicillin-binding protein 1A (Pbp1a) [Chlamydia trachomatis D/UW-3/CX],7U4H_B Chain B, Penicillin-binding protein 1A (Pbp1a) [Chlamydia trachomatis D/UW-3/CX] |
| 3DWK_A | 2.31e-11 | 427 | 587 | 13 | 170 | ChainA, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL],3DWK_B Chain B, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL],3DWK_C Chain C, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL],3DWK_D Chain D, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q478U7 | 1.09e-28 | 419 | 595 | 39 | 213 | Biosynthetic peptidoglycan transglycosylase OS=Dechloromonas aromatica (strain RCB) OX=159087 GN=mtgA PE=3 SV=1 |
| Q7NS41 | 1.38e-28 | 444 | 595 | 61 | 210 | Biosynthetic peptidoglycan transglycosylase OS=Chromobacterium violaceum (strain ATCC 12472 / DSM 30191 / JCM 1249 / NBRC 12614 / NCIMB 9131 / NCTC 9757) OX=243365 GN=mtgA PE=3 SV=1 |
| O24849 | 8.03e-28 | 436 | 590 | 52 | 201 | Biosynthetic peptidoglycan transglycosylase OS=Acinetobacter baylyi (strain ATCC 33305 / BD413 / ADP1) OX=62977 GN=mtgA PE=3 SV=1 |
| Q13U46 | 8.74e-28 | 444 | 587 | 72 | 213 | Biosynthetic peptidoglycan transglycosylase OS=Paraburkholderia xenovorans (strain LB400) OX=266265 GN=mtgA PE=3 SV=1 |
| Q2YBM4 | 1.73e-27 | 446 | 612 | 65 | 224 | Biosynthetic peptidoglycan transglycosylase OS=Nitrosospira multiformis (strain ATCC 25196 / NCIMB 11849 / C 71) OX=323848 GN=mtgA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
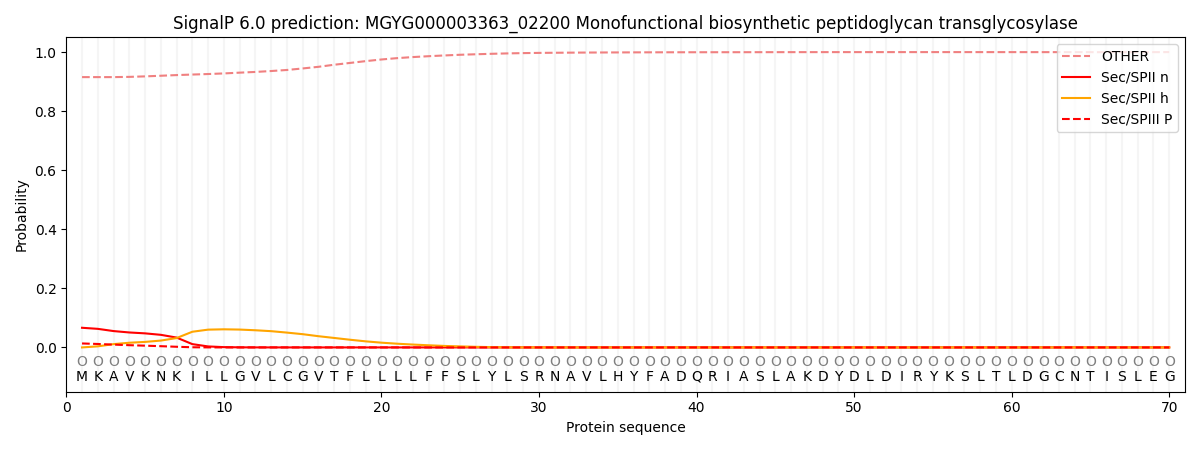
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.915513 | 0.004381 | 0.066572 | 0.000048 | 0.000021 | 0.013490 |