You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003363_02352
You are here: Home > Sequence: MGYG000003363_02352
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides sp900766195 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides sp900766195 | |||||||||||
| CAZyme ID | MGYG000003363_02352 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 44471; End: 46696 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13402 | Peptidase_M60 | 2.52e-48 | 424 | 683 | 1 | 265 | Peptidase M60, enhancin and enhancin-like. This family of peptidases contains a zinc metallopeptidase motif (HEXXHX(8,28)E) and possesses mucinase activity. It includes the viral enhancins as well as enhancin-like peptidases from bacterial species. Enhancins are a class of metalloproteases found in some baculoviruses that enhance viral infection by degrading the peritrophic membrane (PM) of the insect midgut. Bacterial enhancins are found to be cytotoxic when compared to viral enhancin, however, suggesting that the bacterial enhancins do not enhance infection in the same way as viral enhancin. Bacterial enhancins may have evolved a distinct biochemical function. These bacterial domains are peptidases targetting host glycoproteins and thus probably play an important role in successful colonisation of both vertebrate mucosal surfaces and the invertebrate digestive tract by both mutualistic and pathogenic microbes. This family has been augmented by a merge with the sequences in the Enhancin Pfam family. |
| cd14948 | BACON | 2.83e-09 | 33 | 113 | 1 | 82 | Bacteroidetes-Associated Carbohydrate-binding (putative) Often N-terminal (BACON) domain. The BACON domain is found in diverse domain architectures and accociated with a wide variety of domains, including carbohydrate-active enzymes and proteases. It was named for its suggested function of carbohydrate binding; the latter was inferred from domain architectures, sequence conservation, and phyletic distribution. However, recent experimental data suggest that its primary function in Bacteroides ovatus endo-xyloglucanase BoGH5A is to distance the catalytic module from the cell surface and confer additional mobility to the catalytic domain for attack of the polysaccharide. No evidence for a direct role in carbohydrate binding could be found in that case. The large majority of BACON domains are found in Bacteroidetes. |
| pfam00754 | F5_F8_type_C | 1.33e-04 | 151 | 260 | 19 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QCT79445.1 | 1.25e-180 | 34 | 741 | 118 | 860 |
| QRP89238.1 | 1.25e-180 | 34 | 741 | 118 | 860 |
| QUU03487.1 | 1.77e-180 | 34 | 741 | 118 | 860 |
| QRM71921.1 | 3.90e-179 | 34 | 741 | 118 | 860 |
| QTO25368.1 | 3.90e-179 | 34 | 741 | 118 | 860 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7SCI_A | 1.78e-70 | 269 | 734 | 10 | 487 | ChainA, Peptidase M60 domain-containing protein [Akkermansia muciniphila ATCC BAA-835] |
| 5KD2_A | 2.98e-61 | 266 | 740 | 27 | 513 | BT_4244metallopeptidase from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482] |
| 5KD5_A | 3.16e-56 | 314 | 740 | 28 | 465 | BT_4244metallopeptidase from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482],5KD8_A BT_4244 metallopeptidase in complex with Tn antigen. [Bacteroides thetaiotaomicron VPI-5482] |
| 5EV7_A | 4.05e-30 | 345 | 734 | 45 | 407 | Thecrystal structure of a functionally unknown conserved protein mutant from Bacillus anthracis str. Ames [Bacillus anthracis str. Ames] |
| 4FCA_A | 1.80e-28 | 345 | 734 | 45 | 407 | Thecrystal structure of a functionally unknown conserved protein from Bacillus anthracis str. Ames. [Bacillus anthracis str. Ames] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as LIPO
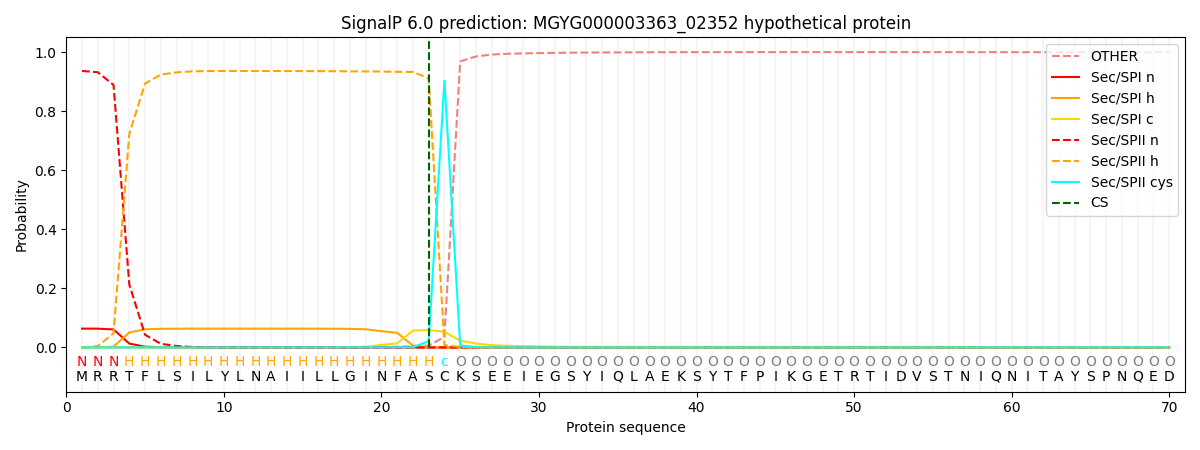
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000439 | 0.061251 | 0.938077 | 0.000079 | 0.000085 | 0.000063 |
