You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003363_03021
You are here: Home > Sequence: MGYG000003363_03021
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides sp900766195 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides sp900766195 | |||||||||||
| CAZyme ID | MGYG000003363_03021 | |||||||||||
| CAZy Family | GH91 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 43842; End: 44987 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH91 | 29 | 374 | 2.3e-88 | 0.9443037974683545 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd21111 | IFTase | 5.58e-51 | 37 | 377 | 42 | 391 | inulin fructotransferase. Inulin fructotransferase (IFTase; EC 4.2.2.17 and EC 4.2.2.18), a member of the glycoside hydrolase family 91, catalyzes depolymerization of beta-2,1-fructans inulin by successively removing the terminal difructosaccharide units as cyclic anhydrides via intramolecular fructosyl transfer. As a result, IFTase produces DFA-I (alpha-D-fructofuranose-beta-D-fructofuranose 2',1:2,1'-dianhydride) and DFA-III (alpha-D-fructofuranose-beta-D-fructofuranose 2',1:2,3'-dianhydride). |
| pfam12708 | Pectate_lyase_3 | 1.81e-06 | 33 | 219 | 4 | 205 | Pectate lyase superfamily protein. This family of proteins possesses a beta helical structure like Pectate lyase. This family is most closely related to glycosyl hydrolase family 28. |
| pfam05048 | NosD | 0.005 | 202 | 323 | 20 | 127 | Periplasmic copper-binding protein (NosD). NosD is a periplasmic protein which is thought to insert copper into the exported reductase apoenzyme (NosZ). This region forms a parallel beta helix domain. |
| COG5434 | Pgu1 | 0.009 | 30 | 72 | 82 | 125 | Polygalacturonase [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QQT79302.1 | 2.94e-123 | 1 | 379 | 2 | 375 |
| QRP58814.1 | 2.94e-123 | 1 | 379 | 2 | 375 |
| ASM68000.1 | 2.94e-123 | 1 | 379 | 2 | 375 |
| QUU10137.1 | 5.95e-121 | 33 | 379 | 32 | 367 |
| QUT25013.1 | 2.37e-119 | 1 | 379 | 1 | 373 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P19870 | 1.23e-14 | 32 | 320 | 8 | 316 | Inulin fructotransferase [DFA-I-forming] OS=Arthrobacter globiformis OX=1665 PE=1 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
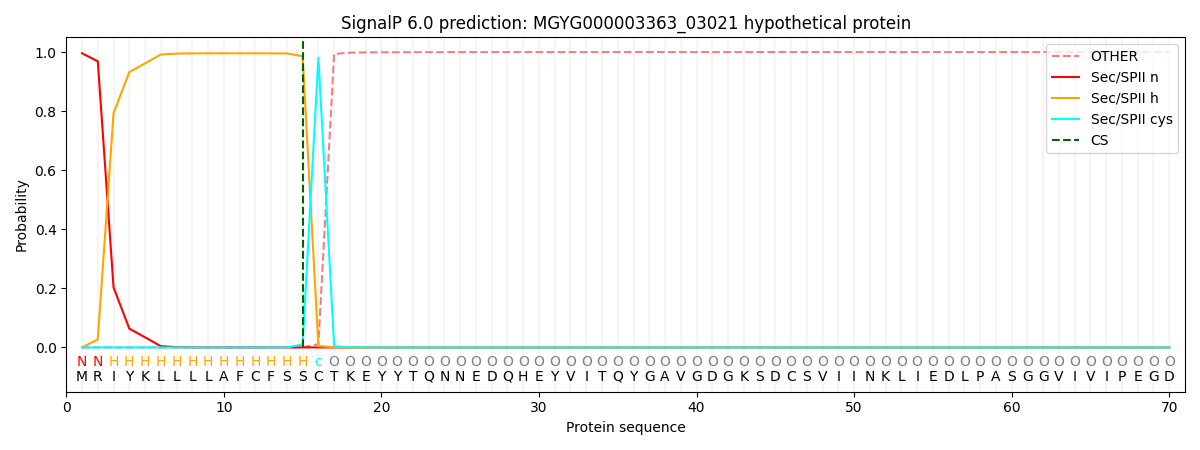
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000029 | 0.003906 | 0.996105 | 0.000003 | 0.000004 | 0.000004 |
