You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003365.1_02559
You are here: Home > Sequence: MGYG000003365.1_02559
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
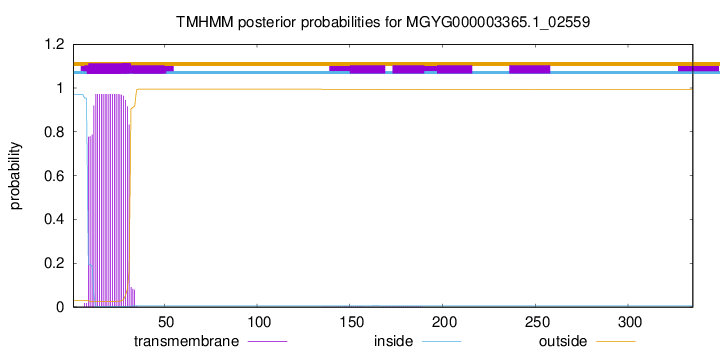
TMHMM annotations
Basic Information help
| Species | Pseudescherichia sp002298805 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Enterobacteriaceae; Pseudescherichia; Pseudescherichia sp002298805 | |||||||||||
| CAZyme ID | MGYG000003365.1_02559 | |||||||||||
| CAZy Family | GH8 | |||||||||||
| CAZyme Description | Endoglucanase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 189067; End: 190074 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01270 | Glyco_hydro_8 | 1.14e-98 | 30 | 334 | 7 | 320 | Glycosyl hydrolases family 8. |
| COG3405 | BcsZ | 1.35e-52 | 12 | 334 | 10 | 345 | Endo-1,4-beta-D-glucanase Y [Carbohydrate transport and metabolism]. |
| PRK11097 | PRK11097 | 3.97e-38 | 8 | 331 | 2 | 352 | cellulase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCU57497.1 | 7.02e-206 | 6 | 334 | 2 | 330 |
| BAY00708.1 | 1.17e-198 | 1 | 335 | 1 | 335 |
| AUV03660.1 | 8.62e-198 | 7 | 335 | 3 | 331 |
| AKL14625.1 | 2.38e-197 | 7 | 335 | 3 | 331 |
| QOV68643.1 | 2.47e-197 | 7 | 335 | 3 | 331 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5CZL_A | 3.63e-174 | 25 | 333 | 26 | 334 | ChainA, Glucanase [Raoultella ornithinolytica] |
| 5GY3_A | 7.64e-167 | 29 | 334 | 2 | 307 | ChainA, Glucanase [Klebsiella pneumoniae] |
| 6VC5_A | 5.42e-69 | 21 | 335 | 1 | 315 | 1.6Angstrom Resolution Crystal Structure of endoglucanase from Komagataeibacter sucrofermentans [Komagataeibacter sucrofermentans] |
| 1WZZ_A | 7.80e-66 | 31 | 332 | 23 | 327 | Structureof endo-beta-1,4-glucanase CMCax from Acetobacter xylinum [Komagataeibacter xylinus] |
| 7F81_A | 2.96e-23 | 30 | 260 | 10 | 242 | ChainA, Glucanase [Enterobacter sp. CJF-002],7F81_B Chain B, Glucanase [Enterobacter sp. CJF-002],7F81_C Chain C, Glucanase [Enterobacter sp. CJF-002],7F81_D Chain D, Glucanase [Enterobacter sp. CJF-002] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P18336 | 2.13e-154 | 8 | 306 | 4 | 302 | Endoglucanase OS=Cellulomonas uda OX=1714 PE=1 SV=1 |
| P27032 | 5.38e-134 | 25 | 332 | 21 | 329 | Minor endoglucanase Y OS=Dickeya dadantii (strain 3937) OX=198628 GN=celY PE=1 SV=1 |
| P37696 | 1.09e-67 | 31 | 332 | 31 | 335 | Probable endoglucanase OS=Komagataeibacter hansenii OX=436 GN=cmcAX PE=1 SV=1 |
| Q8Z289 | 6.60e-25 | 14 | 262 | 12 | 265 | Endoglucanase OS=Salmonella typhi OX=90370 GN=bcsZ PE=3 SV=1 |
| Q8ZLB7 | 2.37e-24 | 15 | 262 | 13 | 265 | Endoglucanase OS=Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) OX=99287 GN=bcsZ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000256 | 0.999005 | 0.000190 | 0.000180 | 0.000167 | 0.000149 |