You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003367_01437
You are here: Home > Sequence: MGYG000003367_01437
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides sp007896885 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides sp007896885 | |||||||||||
| CAZyme ID | MGYG000003367_01437 | |||||||||||
| CAZy Family | GH99 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 251521; End: 252744 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH99 | 50 | 367 | 3.7e-69 | 0.9880239520958084 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd11575 | GH99_GH71_like_3 | 2.11e-151 | 42 | 398 | 1 | 376 | Uncharacterized glycoside hydrolase family 99-like domain. This family of putative glycoside hydrolases resembles glycosyl hydrolase families 71 and 99 (following the CAZY nomenclature) and may share a similar catalytic site and mechanism. |
| cd11573 | GH99_GH71_like | 4.87e-33 | 92 | 354 | 4 | 273 | Glycoside hydrolase families 71, 99, and related domains. This superfamily of glycoside hydrolases contains families GH71 and GH99 (following the CAZY nomenclature), as well as other members with undefined function and specificity. |
| cd11574 | GH99 | 1.33e-22 | 50 | 351 | 1 | 338 | Glycoside hydrolase family 99, an endo-alpha-1,2-mannosidase. This family of glycoside hydrolases 99 (following the CAZY nomenclature) includes endo-alpha-1,2-mannosidase (EC 3.2.1.130), which is an important membrane-associated eukaryotic enzyme involved in the maturation of N-linked glycans. Specifically, it cleaves mannoside linkages internal to N-linked glycan chains by hydrolyzing an alpha-1,2-mannosidic bond between a glucose-substituted mannose and the remainder of the chain. The biological function and significance of the soluble bacterial orthologs, which may have obtained the genes via horizontal transfer, is not clear. |
| pfam16317 | Glyco_hydro_99 | 1.31e-15 | 50 | 338 | 6 | 311 | Glycosyl hydrolase family 99. This domain, around 350 residues, is mainly found in some uncharacterized proteins from bacteroides to human. Some proteins in this family, annotated as endo-alpha-mannosidases cleave mannoside linkages internally within an N-linked glycan chain, short circuiting the classical N-glycan biosynthetic pathway. This domain reveals a (beta-alpha)(8) barrel fold in which the catalytic centre is present in a long substrate-binding groove, consistent with cleavage within the N-glycan chain, providing a foundation upon which to develop new enzyme inhibitors targeting the hijacking of N-glycan synthesis in viral disease and cancer. |
| cd11579 | Glyco_tran_WbsX | 2.55e-14 | 48 | 348 | 1 | 344 | Glycosyl hydrolase family 99-like domain of WbsX-like glycosyltransferases. Members of this domain family are found in proteins within O-antigen biosynthesis clusters in Gram negative bacteria, where they may function as glycosyl hydrolases and typically co-occur with glycosyltransferase domains. They bear resemblance to GH71 and the GH99 family of alpha-1,2-mannosidases and may share a similar cataltyic site and mechanism. The O-antigens are essential lipopolysaccharides in gram-negative bacteria's outer membrane and have been linked to pathogenicity. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUB83810.1 | 2.15e-154 | 14 | 405 | 14 | 406 |
| QKH89593.1 | 1.53e-150 | 1 | 403 | 1 | 404 |
| QUB65436.1 | 2.17e-150 | 1 | 403 | 1 | 404 |
| QUB68154.1 | 6.18e-150 | 15 | 403 | 15 | 404 |
| ASE16633.1 | 1.24e-149 | 15 | 403 | 15 | 404 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4ACZ_A | 1.21e-12 | 54 | 336 | 46 | 340 | Structureof the GH99 endo-alpha-mannosidase from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482],4AD0_A Structure of the GH99 endo-alpha-mannosidase from Bacteriodes thetaiotaomicron in complex with BIS-TRIS-Propane [Bacteroides thetaiotaomicron VPI-5482],4AD0_B Structure of the GH99 endo-alpha-mannosidase from Bacteriodes thetaiotaomicron in complex with BIS-TRIS-Propane [Bacteroides thetaiotaomicron VPI-5482],4AD0_C Structure of the GH99 endo-alpha-mannosidase from Bacteriodes thetaiotaomicron in complex with BIS-TRIS-Propane [Bacteroides thetaiotaomicron VPI-5482],4AD0_D Structure of the GH99 endo-alpha-mannosidase from Bacteriodes thetaiotaomicron in complex with BIS-TRIS-Propane [Bacteroides thetaiotaomicron VPI-5482] |
| 4ACZ_B | 1.61e-12 | 54 | 336 | 46 | 340 | Structureof the GH99 endo-alpha-mannosidase from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482] |
| 4ACY_A | 2.88e-12 | 54 | 336 | 46 | 340 | Selenomethioninederivative of the GH99 endo-alpha-mannosidase from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482],4ACY_B Selenomethionine derivative of the GH99 endo-alpha-mannosidase from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482] |
| 4AD1_A | 3.82e-09 | 51 | 355 | 42 | 357 | Structureof the GH99 endo-alpha-mannosidase from Bacteroides xylanisolvens [Bacteroides xylanisolvens XB1A],4AD2_A Structure of the GH99 endo-alpha-mannosidase from Bacteroides xylanisolvens in complex with glucose-1,3-isofagomine [Bacteroides xylanisolvens XB1A],4AD2_B Structure of the GH99 endo-alpha-mannosidase from Bacteroides xylanisolvens in complex with glucose-1,3-isofagomine [Bacteroides xylanisolvens XB1A],4AD2_C Structure of the GH99 endo-alpha-mannosidase from Bacteroides xylanisolvens in complex with glucose-1,3-isofagomine [Bacteroides xylanisolvens XB1A],4AD2_D Structure of the GH99 endo-alpha-mannosidase from Bacteroides xylanisolvens in complex with glucose-1,3-isofagomine [Bacteroides xylanisolvens XB1A],4AD3_A Structure of the GH99 endo-alpha-mannosidase from Bacteroides xylanisolvens in complex with Glucose-1,3-deoxymannojirimycin [Bacteroides xylanisolvens XB1A],4AD3_B Structure of the GH99 endo-alpha-mannosidase from Bacteroides xylanisolvens in complex with Glucose-1,3-deoxymannojirimycin [Bacteroides xylanisolvens XB1A],4AD3_C Structure of the GH99 endo-alpha-mannosidase from Bacteroides xylanisolvens in complex with Glucose-1,3-deoxymannojirimycin [Bacteroides xylanisolvens XB1A],4AD3_D Structure of the GH99 endo-alpha-mannosidase from Bacteroides xylanisolvens in complex with Glucose-1,3-deoxymannojirimycin [Bacteroides xylanisolvens XB1A],4AD4_A Structure of the GH99 endo-alpha-mannosidase from Bacteroides xylanisolvens in complex with glucose-1,3-isofagomine and alpha-1,2- mannobiose [Bacteroides xylanisolvens XB1A],4AD5_A Structure of the GH99 endo-alpha-mannosidase from Bacteroides xylanisolvens in complex with glucose-1,3-deoxymannojirimycin and alpha-1,2-mannobiose [Bacteroides xylanisolvens XB1A],4UTF_A Structure of the GH99 endo-alpha-mannosidase from Bacteroides xylanisolvens in complex with mannose-alpha-1,3-isofagomine and alpha- 1,2-mannobiose [Bacteroides xylanisolvens XB1A],4V27_A Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with mannose-alpha-1,3-isofagomine [Bacteroides xylanisolvens XB1A] |
| 5LYR_A | 3.88e-09 | 51 | 355 | 47 | 362 | Structureof the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with mannose-alpha-1,3-noeuromycin [Bacteroides xylanisolvens XB1A],5M03_A Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with mannose-alpha-1,3-noeuromycin and 1,2-alpha-mannobiose [Bacteroides xylanisolvens XB1A],5M17_A Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with mannose-alpha-1,3-1,2-dideoxymannose [Bacteroides xylanisolvens XB1A],5M3W_A Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with mannose-alpha-1,3-1,2-dideoxymannose and alpha-1,2-mannobiose [Bacteroides xylanisolvens XB1A],5M5D_A Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with mannose-alpha-1,3-D-glucal [Bacteroides xylanisolvens XB1A],5MC8_A Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with mannose-alpha-1,3-D-glucal and alpha-1,2-mannobiose [Bacteroides xylanisolvens XB1A],6FAM_A Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with mannose-alpha-1,3-2-aminodeoxymannojirimycin [Bacteroides xylanisolvens XB1A],6FAR_A Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with mannose-alpha-1,3-mannoimidazole [Bacteroides xylanisolvens XB1A],6FWI_A Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with alpha-Glc-1,3-(1,2-anhydro-carba-mannosamine) [Bacteroides xylanisolvens XB1A],6FWJ_A Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with alpha-Glc-1,3-(1,2-anhydro-carba-mannosamine) and alpha-1,2-mannobiose [Bacteroides xylanisolvens XB1A],6FWM_A Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with alpha-Glc-1,3-1,2-anhydro-mannose hydrolyzed by enzyme [Bacteroides xylanisolvens XB1A],6FWP_A Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with alpha-1,3-mannobiose and alpha-1,2-mannobiose [Bacteroides xylanisolvens XB1A],6HMG_A Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with alpha-Glc-1,3-(1,2-anhydro-carba-glucosamine) [Bacteroides xylanisolvens XB1A],6HMH_A Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with alpha-Glc-1,3-(1,2-anhydro-carba-glucosamine) and alpha-1,2-mannobiose [Bacteroides xylanisolvens XB1A],6ZJ6_AAA Chain AAA, Glycosyl hydrolase family 71 [Bacteroides xylanisolvens XB1A] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q6P1J0 | 8.41e-11 | 34 | 365 | 83 | 450 | Glycoprotein endo-alpha-1,2-mannosidase-like protein OS=Mus musculus OX=10090 GN=Maneal PE=2 SV=1 |
| Q5VSG8 | 4.72e-10 | 39 | 336 | 93 | 417 | Glycoprotein endo-alpha-1,2-mannosidase-like protein OS=Homo sapiens OX=9606 GN=MANEAL PE=2 SV=1 |
| Q1L8D2 | 1.02e-07 | 50 | 333 | 90 | 399 | Glycoprotein endo-alpha-1,2-mannosidase-like protein OS=Danio rerio OX=7955 GN=maneal PE=3 SV=1 |
| Q6NXH2 | 1.01e-06 | 21 | 336 | 72 | 415 | Glycoprotein endo-alpha-1,2-mannosidase OS=Mus musculus OX=10090 GN=Manea PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
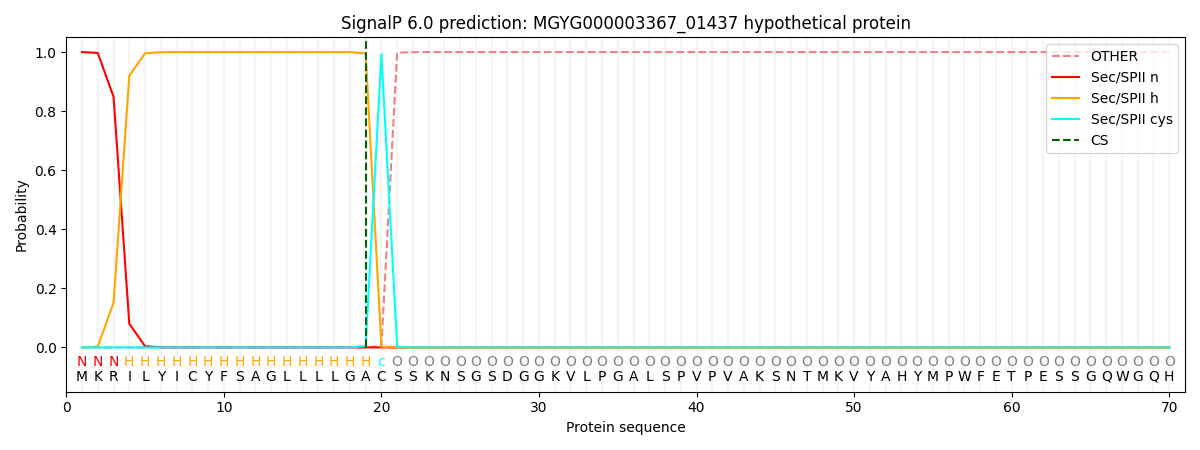
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000045 | 0.000000 | 0.000000 | 0.000000 |
