You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003380_00786
You are here: Home > Sequence: MGYG000003380_00786
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Corynebacterium aurimucosum_E | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Mycobacteriales; Mycobacteriaceae; Corynebacterium; Corynebacterium aurimucosum_E | |||||||||||
| CAZyme ID | MGYG000003380_00786 | |||||||||||
| CAZy Family | CBM48 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 70250; End: 73663 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1674 | FtsK | 3.68e-158 | 581 | 1069 | 368 | 858 | DNA segregation ATPase FtsK/SpoIIIE and related proteins [Cell cycle control, cell division, chromosome partitioning]. |
| PRK10263 | PRK10263 | 3.49e-145 | 596 | 1066 | 866 | 1352 | DNA translocase FtsK; Provisional |
| pfam01580 | FtsK_SpoIIIE | 2.15e-64 | 702 | 901 | 1 | 219 | FtsK/SpoIIIE family. FtsK has extensive sequence similarity to wide variety of proteins from prokaryotes and plasmids, termed the FtsK/SpoIIIE family. This domain contains a putative ATP binding P-loop motif. It is found in the FtsK cell division protein from E. coli and the stage III sporulation protein E SpoIIIE which has roles in regulation of prespore specific gene expression in B. subtilis. A mutation in FtsK causes a temperature sensitive block in cell division and it is involved in peptidoglycan synthesis or modification. The SpoIIIE protein is implicated in intercellular chromosomal DNA transfer. |
| pfam17854 | FtsK_alpha | 2.52e-34 | 597 | 694 | 2 | 101 | FtsK alpha domain. FtsK is a DNA translocase that coordinates chromosome segregation and cell division in bacteria. In addition to its role as activator of XerCD site-specific recombination, FtsK can translocate double-stranded DNA (dsDNA) rapidly and directionally and reverse direction. FtsK can be split into three domains called alpha (this entry), beta and gamma. The alpha and beta domains contain the core ATPase machinery of the DNA translocase. |
| pfam09397 | Ftsk_gamma | 4.52e-27 | 1005 | 1065 | 2 | 63 | Ftsk gamma domain. This domain directs oriented DNA translocation and forms a winged helix structure. Mutated proteins with substitutions in the FtsK gamma DNA-recognition helix are impaired in DNA binding. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AGL63871.2 | 1.07e-117 | 594 | 1069 | 526 | 989 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2IUS_A | 8.61e-129 | 596 | 1066 | 23 | 509 | E.coli FtsK motor domain [Escherichia coli],2IUS_B E. coli FtsK motor domain [Escherichia coli],2IUS_C E. coli FtsK motor domain [Escherichia coli],2IUS_D E. coli FtsK motor domain [Escherichia coli],2IUS_E E. coli FtsK motor domain [Escherichia coli],2IUS_F E. coli FtsK motor domain [Escherichia coli] |
| 2IUT_A | 3.91e-122 | 582 | 1063 | 45 | 559 | P.aeruginosa FtsK motor domain, dimeric [Pseudomonas aeruginosa],2IUT_B P. aeruginosa FtsK motor domain, dimeric [Pseudomonas aeruginosa] |
| 2IUU_A | 4.24e-113 | 582 | 986 | 45 | 475 | P.aeruginosa FtsK motor domain, hexamer [Pseudomonas aeruginosa],2IUU_B P. aeruginosa FtsK motor domain, hexamer [Pseudomonas aeruginosa],2IUU_C P. aeruginosa FtsK motor domain, hexamer [Pseudomonas aeruginosa],2IUU_D P. aeruginosa FtsK motor domain, hexamer [Pseudomonas aeruginosa],2IUU_E P. aeruginosa FtsK motor domain, hexamer [Pseudomonas aeruginosa],2IUU_F P. aeruginosa FtsK motor domain, hexamer [Pseudomonas aeruginosa],6T8B_A FtsK motor domain with dsDNA, translocating state [Pseudomonas aeruginosa PAO1],6T8B_B FtsK motor domain with dsDNA, translocating state [Pseudomonas aeruginosa PAO1],6T8B_C FtsK motor domain with dsDNA, translocating state [Pseudomonas aeruginosa PAO1],6T8B_D FtsK motor domain with dsDNA, translocating state [Pseudomonas aeruginosa PAO1],6T8B_E FtsK motor domain with dsDNA, translocating state [Pseudomonas aeruginosa PAO1],6T8B_F FtsK motor domain with dsDNA, translocating state [Pseudomonas aeruginosa PAO1],6T8G_A Stalled FtsK motor domain bound to dsDNA [Pseudomonas aeruginosa PAO1],6T8G_B Stalled FtsK motor domain bound to dsDNA [Pseudomonas aeruginosa PAO1],6T8G_C Stalled FtsK motor domain bound to dsDNA [Pseudomonas aeruginosa PAO1],6T8G_D Stalled FtsK motor domain bound to dsDNA [Pseudomonas aeruginosa PAO1],6T8G_E Stalled FtsK motor domain bound to dsDNA [Pseudomonas aeruginosa PAO1],6T8G_F Stalled FtsK motor domain bound to dsDNA [Pseudomonas aeruginosa PAO1],6T8O_A Stalled FtsK motor domain bound to dsDNA end [Pseudomonas aeruginosa PAO1],6T8O_B Stalled FtsK motor domain bound to dsDNA end [Pseudomonas aeruginosa PAO1],6T8O_C Stalled FtsK motor domain bound to dsDNA end [Pseudomonas aeruginosa PAO1],6T8O_D Stalled FtsK motor domain bound to dsDNA end [Pseudomonas aeruginosa PAO1],6T8O_E Stalled FtsK motor domain bound to dsDNA end [Pseudomonas aeruginosa PAO1],6T8O_F Stalled FtsK motor domain bound to dsDNA end [Pseudomonas aeruginosa PAO1] |
| 6O1X_A | 7.10e-15 | 719 | 966 | 111 | 347 | Structureof pCW3 conjugation coupling protein TcpA monomer form with ATPgS [Clostridium perfringens],6O1X_B Structure of pCW3 conjugation coupling protein TcpA monomer form with ATPgS [Clostridium perfringens],6O1Y_A Structure of pCW3 conjugation coupling protein TcpA monomeric form with ATP [Clostridium perfringens],6O1Y_B Structure of pCW3 conjugation coupling protein TcpA monomeric form with ATP [Clostridium perfringens] |
| 6O1Z_A | 7.52e-15 | 719 | 966 | 111 | 347 | Structureof pCW3 conjugation coupling protein TcpA hexagonal crystal form [Clostridium perfringens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8FPC1 | 0.0 | 148 | 1137 | 74 | 946 | DNA translocase FtsK OS=Corynebacterium efficiens (strain DSM 44549 / YS-314 / AJ 12310 / JCM 11189 / NBRC 100395) OX=196164 GN=ftsK PE=3 SV=2 |
| Q8NP53 | 5.48e-314 | 81 | 1087 | 4 | 892 | DNA translocase FtsK OS=Corynebacterium glutamicum (strain ATCC 13032 / DSM 20300 / BCRC 11384 / JCM 1318 / LMG 3730 / NCIMB 10025) OX=196627 GN=ftsK PE=3 SV=1 |
| O05560 | 5.21e-247 | 151 | 1087 | 90 | 839 | DNA translocase FtsK OS=Mycobacterium leprae (strain TN) OX=272631 GN=ftsK PE=3 SV=2 |
| P9WNA3 | 3.07e-246 | 151 | 1078 | 96 | 827 | DNA translocase FtsK OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=ftsK PE=1 SV=1 |
| P9WNA2 | 3.07e-246 | 151 | 1078 | 96 | 827 | DNA translocase FtsK OS=Mycobacterium tuberculosis (strain CDC 1551 / Oshkosh) OX=83331 GN=ftsK PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.997258 | 0.002709 | 0.000018 | 0.000009 | 0.000006 | 0.000007 |
TMHMM Annotations download full data without filtering help
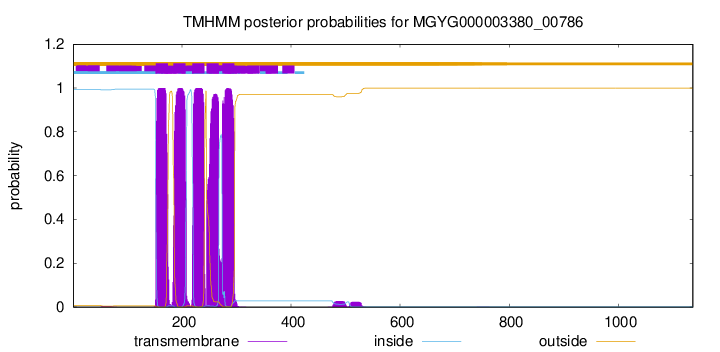
| start | end |
|---|---|
| 152 | 174 |
| 184 | 206 |
| 219 | 241 |
| 245 | 267 |
| 274 | 296 |
