You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003383_02421
You are here: Home > Sequence: MGYG000003383_02421
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
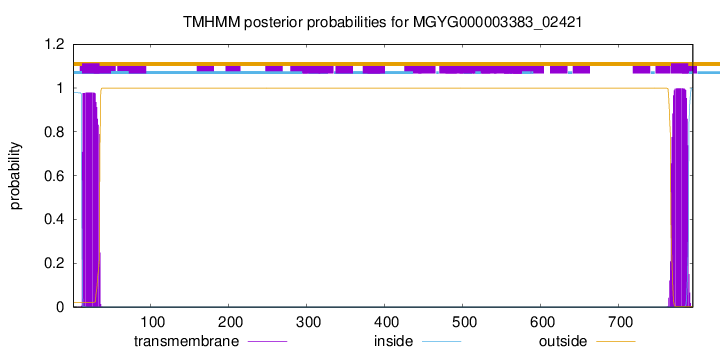
TMHMM annotations
Basic Information help
| Species | Bifidobacterium scardovii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Bifidobacteriaceae; Bifidobacterium; Bifidobacterium scardovii | |||||||||||
| CAZyme ID | MGYG000003383_02421 | |||||||||||
| CAZy Family | GH26 | |||||||||||
| CAZyme Description | Mannan endo-1,4-beta-mannosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 69393; End: 71780 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH26 | 53 | 371 | 5.2e-61 | 0.9108910891089109 |
| CBM23 | 519 | 682 | 2.7e-36 | 0.9876543209876543 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02156 | Glyco_hydro_26 | 1.23e-43 | 53 | 354 | 2 | 299 | Glycosyl hydrolase family 26. |
| COG4124 | ManB2 | 1.15e-18 | 153 | 331 | 117 | 299 | Beta-mannanase [Carbohydrate transport and metabolism]. |
| pfam03425 | CBM_11 | 4.71e-08 | 520 | 682 | 7 | 172 | Carbohydrate binding domain (family 11). |
| pfam04886 | PT | 3.77e-04 | 714 | 744 | 1 | 33 | PT repeat. This short repeat is composed on the tetrapeptide XPTX. This repeat is found in a variety of proteins, however it is not clear if these repeats are homologous to each other. The alignment represents nine copies of this repeat. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BAQ30247.1 | 0.0 | 1 | 795 | 1 | 795 |
| ASR56276.1 | 1.86e-163 | 10 | 687 | 11 | 694 |
| CQR64988.1 | 1.76e-160 | 5 | 680 | 4 | 668 |
| QTL79031.1 | 1.54e-159 | 47 | 687 | 38 | 703 |
| AEI10833.1 | 2.51e-159 | 20 | 687 | 28 | 700 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2BVT_A | 6.23e-105 | 48 | 501 | 5 | 458 | Thestructure of a modular endo-beta-1,4-mannanase from Cellulomonas fimi explains the product specificity of glycoside hydrolase family 26 mannanases. [Cellulomonas fimi],2BVT_B The structure of a modular endo-beta-1,4-mannanase from Cellulomonas fimi explains the product specificity of glycoside hydrolase family 26 mannanases. [Cellulomonas fimi],2BVY_A The structure and characterization of a modular endo-beta-1,4-mannanase from Cellulomonas fimi [Cellulomonas fimi] |
| 2X2Y_A | 5.11e-103 | 48 | 501 | 5 | 458 | Cellulomonasfimi endo-beta-1,4-mannanase double mutant [Cellulomonas fimi],2X2Y_B Cellulomonas fimi endo-beta-1,4-mannanase double mutant [Cellulomonas fimi] |
| 3TP4_A | 1.94e-91 | 48 | 501 | 5 | 458 | CrystalStructure of engineered protein at the resolution 1.98A, Northeast Structural Genomics Consortium Target OR128 [synthetic construct],3TP4_B Crystal Structure of engineered protein at the resolution 1.98A, Northeast Structural Genomics Consortium Target OR128 [synthetic construct] |
| 4YN5_A | 3.91e-84 | 51 | 380 | 51 | 387 | Catalyticdomain of Bacillus sp. JAMB-750 GH26 Endo-beta-1,4-mannanase [Bacillus sp. JAMB750] |
| 1J9Y_A | 1.49e-70 | 48 | 411 | 8 | 362 | Crystalstructure of mannanase 26A from Pseudomonas cellulosa [Cellvibrio japonicus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A1A278 | 1.21e-151 | 47 | 687 | 38 | 703 | Mannan endo-1,4-beta-mannosidase OS=Bifidobacterium adolescentis (strain ATCC 15703 / DSM 20083 / NCTC 11814 / E194a) OX=367928 GN=BAD_1030 PE=1 SV=1 |
| P49424 | 2.34e-69 | 48 | 411 | 46 | 400 | Mannan endo-1,4-beta-mannosidase OS=Cellvibrio japonicus (strain Ueda107) OX=498211 GN=manA PE=1 SV=2 |
| O05512 | 9.23e-18 | 99 | 294 | 79 | 272 | Mannan endo-1,4-beta-mannosidase OS=Bacillus subtilis (strain 168) OX=224308 GN=gmuG PE=1 SV=2 |
| P55278 | 7.33e-16 | 40 | 293 | 18 | 269 | Mannan endo-1,4-beta-mannosidase OS=Bacillus subtilis OX=1423 PE=3 SV=1 |
| Q5AWB7 | 7.41e-14 | 82 | 367 | 82 | 319 | Probable mannan endo-1,4-beta-mannosidase E OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=manE PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
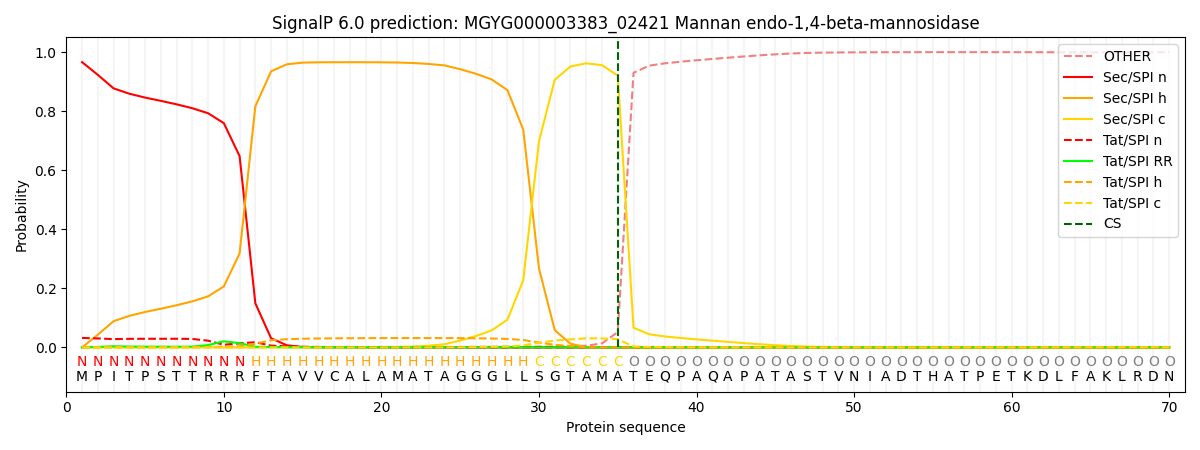
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.002356 | 0.962809 | 0.000801 | 0.033383 | 0.000404 | 0.000208 |