You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003387_03269
You are here: Home > Sequence: MGYG000003387_03269
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Pseudomonas_R sp900766265 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Pseudomonadales; Pseudomonadaceae; Pseudomonas_R; Pseudomonas_R sp900766265 | |||||||||||
| CAZyme ID | MGYG000003387_03269 | |||||||||||
| CAZy Family | GT0 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 738; End: 3740 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0744 | MrcB | 2.53e-60 | 60 | 922 | 5 | 661 | Membrane carboxypeptidase (penicillin-binding protein) [Cell wall/membrane/envelope biogenesis]. |
| pfam00912 | Transgly | 3.22e-21 | 121 | 327 | 1 | 172 | Transglycosylase. The penicillin-binding proteins are bifunctional proteins consisting of transglycosylase and transpeptidase in the N- and C-terminus respectively. The transglycosylase domain catalyzes the polymerization of murein glycan chains. |
| COG0768 | FtsI | 1.11e-05 | 817 | 909 | 444 | 528 | Cell division protein FtsI/penicillin-binding protein 2 [Cell cycle control, cell division, chromosome partitioning, Cell wall/membrane/envelope biogenesis]. |
| COG4953 | PbpC | 1.12e-04 | 119 | 422 | 41 | 292 | Membrane carboxypeptidase/penicillin-binding protein PbpC [Cell wall/membrane/envelope biogenesis]. |
| pfam00905 | Transpeptidase | 4.97e-04 | 817 | 910 | 161 | 237 | Penicillin binding protein transpeptidase domain. The active site serine is conserved in all members of this family. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AEA83854.1 | 0.0 | 11 | 991 | 3 | 982 |
| AEJ05108.1 | 0.0 | 11 | 991 | 3 | 982 |
| ABP79605.1 | 0.0 | 13 | 991 | 1 | 978 |
| AEB57320.1 | 0.0 | 8 | 998 | 33 | 1023 |
| ACO77613.1 | 0.0 | 4 | 994 | 31 | 1026 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
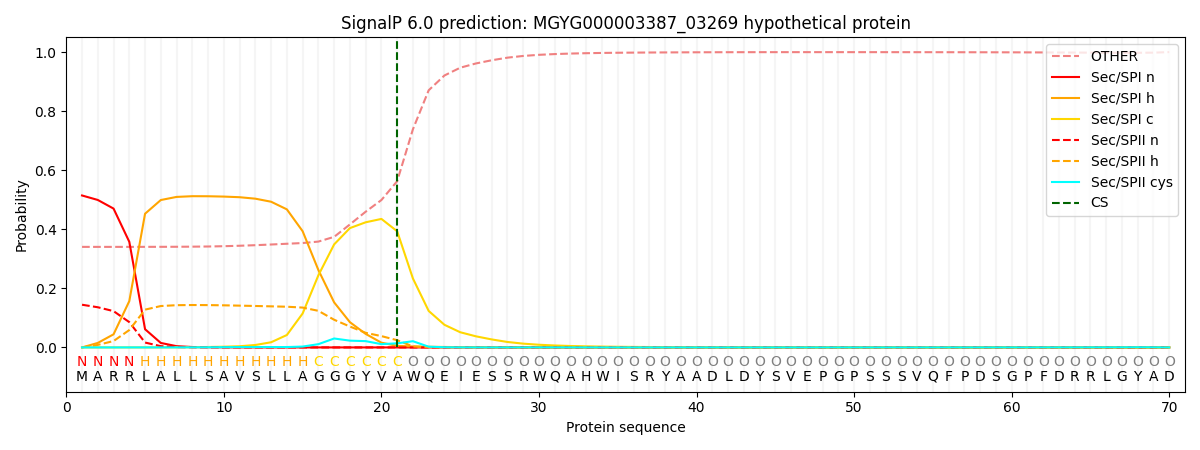
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.351360 | 0.500134 | 0.146621 | 0.000776 | 0.000482 | 0.000603 |
