You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003393_01627
You are here: Home > Sequence: MGYG000003393_01627
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Pantoea ananatis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Enterobacteriaceae; Pantoea; Pantoea ananatis | |||||||||||
| CAZyme ID | MGYG000003393_01627 | |||||||||||
| CAZy Family | GT83 | |||||||||||
| CAZyme Description | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 161308; End: 163047 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT83 | 47 | 522 | 3.1e-55 | 0.8666666666666667 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1807 | ArnT | 4.94e-26 | 49 | 530 | 9 | 476 | 4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| pfam13231 | PMT_2 | 1.45e-10 | 104 | 264 | 2 | 160 | Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| UAC69887.1 | 3.91e-55 | 52 | 575 | 63 | 621 |
| ACB74694.1 | 9.00e-15 | 42 | 206 | 5 | 172 |
| QDU20239.1 | 4.01e-13 | 47 | 180 | 31 | 161 |
| AWI08836.1 | 1.19e-12 | 55 | 199 | 113 | 254 |
| AVL27167.1 | 3.19e-12 | 54 | 493 | 21 | 458 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A8GDR9 | 1.07e-08 | 49 | 405 | 9 | 372 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Serratia proteamaculans (strain 568) OX=399741 GN=arnT PE=3 SV=1 |
| Q4ZSZ0 | 2.76e-06 | 50 | 377 | 9 | 335 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Pseudomonas syringae pv. syringae (strain B728a) OX=205918 GN=arnT PE=3 SV=1 |
| C5BDQ8 | 3.64e-06 | 55 | 378 | 12 | 336 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Edwardsiella ictaluri (strain 93-146) OX=634503 GN=arnT PE=3 SV=1 |
| A1JPM4 | 8.38e-06 | 51 | 377 | 11 | 340 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Yersinia enterocolitica serotype O:8 / biotype 1B (strain NCTC 13174 / 8081) OX=393305 GN=arnT PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.999597 | 0.000413 | 0.000001 | 0.000000 | 0.000000 | 0.000001 |
TMHMM Annotations download full data without filtering help
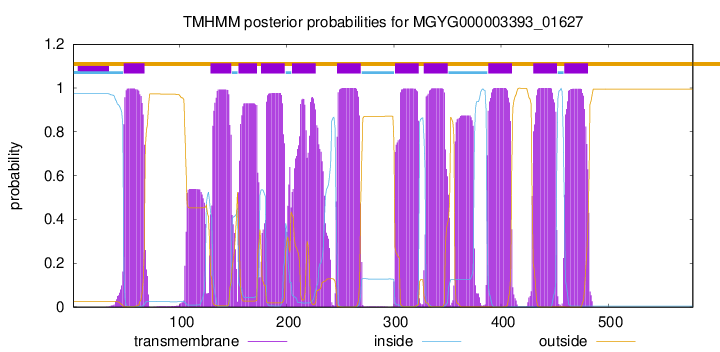
| start | end |
|---|---|
| 48 | 67 |
| 129 | 148 |
| 155 | 172 |
| 176 | 198 |
| 205 | 227 |
| 247 | 269 |
| 301 | 323 |
| 328 | 350 |
| 388 | 410 |
| 430 | 452 |
| 459 | 481 |
