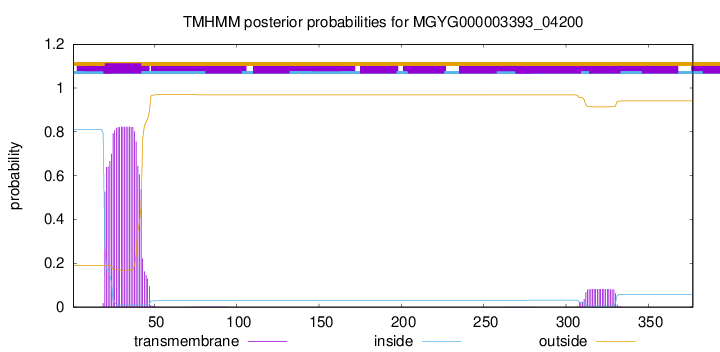You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003393_04200
You are here: Home > Sequence: MGYG000003393_04200
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Pantoea ananatis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Enterobacteriaceae; Pantoea; Pantoea ananatis | |||||||||||
| CAZyme ID | MGYG000003393_04200 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 170454; End: 171587 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 47 | 368 | 2.7e-81 | 0.9900990099009901 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00331 | Glyco_hydro_10 | 1.68e-94 | 47 | 365 | 1 | 307 | Glycosyl hydrolase family 10. |
| smart00633 | Glyco_10 | 4.23e-82 | 90 | 365 | 1 | 262 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 3.78e-74 | 29 | 365 | 6 | 336 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QKV89082.1 | 3.26e-278 | 1 | 377 | 35 | 411 |
| CCF11431.1 | 1.89e-277 | 1 | 377 | 35 | 411 |
| QAB32087.1 | 1.89e-277 | 1 | 377 | 35 | 411 |
| ADD75543.1 | 1.89e-277 | 1 | 377 | 35 | 411 |
| QTC45991.1 | 3.81e-277 | 1 | 377 | 35 | 411 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6FHE_A | 5.52e-51 | 59 | 365 | 24 | 337 | Highlyactive enzymes by automated modular backbone assembly and sequence design [synthetic construct] |
| 1NQ6_A | 3.68e-48 | 46 | 370 | 3 | 302 | CrystalStructure of the catalytic domain of xylanase A from Streptomyces halstedii JM8 [Streptomyces halstedii] |
| 2G3I_A | 3.74e-47 | 47 | 370 | 5 | 300 | Structureof S.olivaceoviridis xylanase Q88A/R275A mutant [Streptomyces olivaceoviridis],2G3J_A Structure of S.olivaceoviridis xylanase Q88A/R275A mutant [Streptomyces olivaceoviridis],2G4F_A Chain A, Hydrolase [Streptomyces olivaceoviridis],2G4F_B Chain B, Hydrolase [Streptomyces olivaceoviridis] |
| 5M0K_A | 4.31e-47 | 12 | 372 | 3 | 337 | CRYSTALSTRUCTURE of endo-1,4-beta-xylanase from Cellulomonas flavigena [Cellulomonas flavigena],5M0K_B CRYSTAL STRUCTURE of endo-1,4-beta-xylanase from Cellulomonas flavigena [Cellulomonas flavigena] |
| 1EXP_A | 7.17e-47 | 47 | 331 | 4 | 277 | Beta-1,4-GlycanaseCex-Cd [Cellulomonas fimi],1FH7_A Crystal Structure Of The Xylanase Cex With Xylobiose- Derived Inhibitor Deoxynojirimycin [Cellulomonas fimi],1FH8_A Crystal Structure Of The Xylanase Cex With Xylobiose-derived Isofagomine Inhibitor [Cellulomonas fimi],1FH9_A Crystal Structure Of The Xylanase Cex With Xylobiose-derived Lactam Oxime Inhibitor [Cellulomonas fimi],1FHD_A Crystal Structure Of The Xylanase Cex With Xylobiose-derived Imidazole Inhibitor [Cellulomonas fimi],1J01_A Crystal Structure Of The Xylanase Cex With Xylobiose-Derived Inhibitor Isofagomine lactam [Cellulomonas fimi],2EXO_A Crystal Structure Of The Catalytic Domain Of The Beta-1,4- Glycanase Cex From Cellulomonas Fimi [Cellulomonas fimi],2XYL_A Cellulomonas Fimi XylanaseCELLULASE COMPLEXED WITH 2-Deoxy- 2-Fluoro-Xylobiose [Cellulomonas fimi] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P07986 | 4.08e-47 | 11 | 374 | 17 | 357 | Exoglucanase/xylanase OS=Cellulomonas fimi OX=1708 GN=cex PE=1 SV=1 |
| O94163 | 1.58e-45 | 52 | 370 | 36 | 327 | Endo-1,4-beta-xylanase F1 OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=xynF1 PE=1 SV=1 |
| Q4P902 | 1.71e-45 | 50 | 372 | 45 | 341 | Endo-1,4-beta-xylanase UM03411 OS=Ustilago maydis (strain 521 / FGSC 9021) OX=237631 GN=UMAG_03411 PE=1 SV=1 |
| P56588 | 1.25e-44 | 84 | 370 | 44 | 302 | Endo-1,4-beta-xylanase OS=Penicillium simplicissimum OX=69488 PE=1 SV=1 |
| Q5S7A8 | 2.35e-44 | 84 | 370 | 69 | 327 | Endo-1,4-beta-xylanase A OS=Penicillium canescens OX=5083 GN=xylA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as TAT
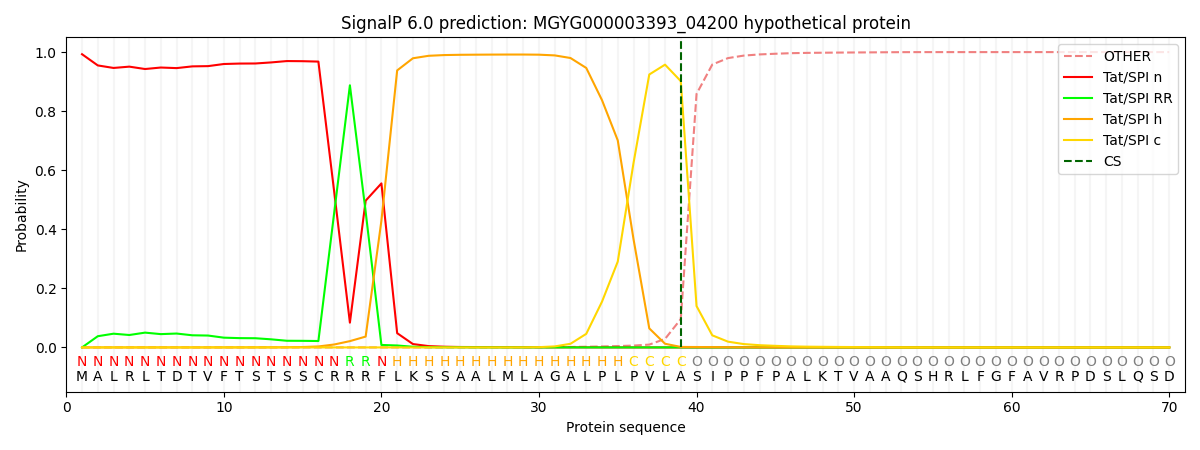
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000009 | 0.000017 | 0.000003 | 0.992818 | 0.007142 | 0.000000 |