You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003396_00305
You are here: Home > Sequence: MGYG000003396_00305
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Clostridium sp900766315 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Clostridiales; Clostridiaceae; Clostridium; Clostridium sp900766315 | |||||||||||
| CAZyme ID | MGYG000003396_00305 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 17526; End: 24884 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM51 | 1831 | 1970 | 1.6e-44 | 0.9850746268656716 |
| CBM51 | 2056 | 2196 | 1.9e-43 | 0.9925373134328358 |
| CBM51 | 1312 | 1446 | 3.9e-32 | 0.9850746268656716 |
| CBM32 | 80 | 206 | 6.7e-16 | 0.9354838709677419 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam08305 | NPCBM | 9.58e-51 | 2054 | 2197 | 1 | 136 | NPCBM/NEW2 domain. This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. This domain has also been called the NEW2 domain (Naumoff DG. Phylogenetic analysis of alpha-galactosidases of the GH27 family. Molecular Biology (Engl Transl). (2004)38:388-399.) |
| pfam08305 | NPCBM | 2.21e-49 | 1832 | 1970 | 5 | 135 | NPCBM/NEW2 domain. This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. This domain has also been called the NEW2 domain (Naumoff DG. Phylogenetic analysis of alpha-galactosidases of the GH27 family. Molecular Biology (Engl Transl). (2004)38:388-399.) |
| smart00776 | NPCBM | 2.19e-45 | 1827 | 1970 | 2 | 144 | This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. |
| smart00776 | NPCBM | 3.68e-44 | 2056 | 2197 | 5 | 145 | This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. |
| pfam08305 | NPCBM | 6.93e-40 | 1309 | 1447 | 1 | 136 | NPCBM/NEW2 domain. This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. This domain has also been called the NEW2 domain (Naumoff DG. Phylogenetic analysis of alpha-galactosidases of the GH27 family. Molecular Biology (Engl Transl). (2004)38:388-399.) |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QJS19250.1 | 0.0 | 2 | 2450 | 1 | 2288 |
| AQM59432.1 | 0.0 | 3 | 2452 | 3 | 2445 |
| QPQ34479.1 | 0.0 | 17 | 2451 | 15 | 1934 |
| AHN23111.1 | 0.0 | 17 | 2439 | 15 | 1921 |
| AVK95662.1 | 0.0 | 17 | 2439 | 15 | 1921 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7JFS_A | 1.08e-178 | 45 | 1230 | 22 | 981 | ChainA, F5/8 type C domain protein [Clostridium perfringens ATCC 13124] |
| 7JS4_A | 1.14e-128 | 746 | 1525 | 49 | 831 | ChainA, F5/8 type C domain protein [Clostridium perfringens ATCC 13124] |
| 7JND_A | 2.54e-118 | 37 | 454 | 29 | 449 | ChainA, F5/8 type C domain protein [Clostridium perfringens ATCC 13124],7JNF_A Chain A, F5/8 type C domain protein [Clostridium perfringens ATCC 13124] |
| 7JNB_A | 1.02e-116 | 37 | 454 | 29 | 449 | ChainA, F5/8 type C domain protein [Clostridium perfringens ATCC 13124] |
| 5KDJ_A | 1.40e-94 | 656 | 1308 | 24 | 672 | ZmpBmetallopeptidase from Clostridium perfringens [Clostridium perfringens ATCC 13124],5KDJ_B ZmpB metallopeptidase from Clostridium perfringens [Clostridium perfringens ATCC 13124] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
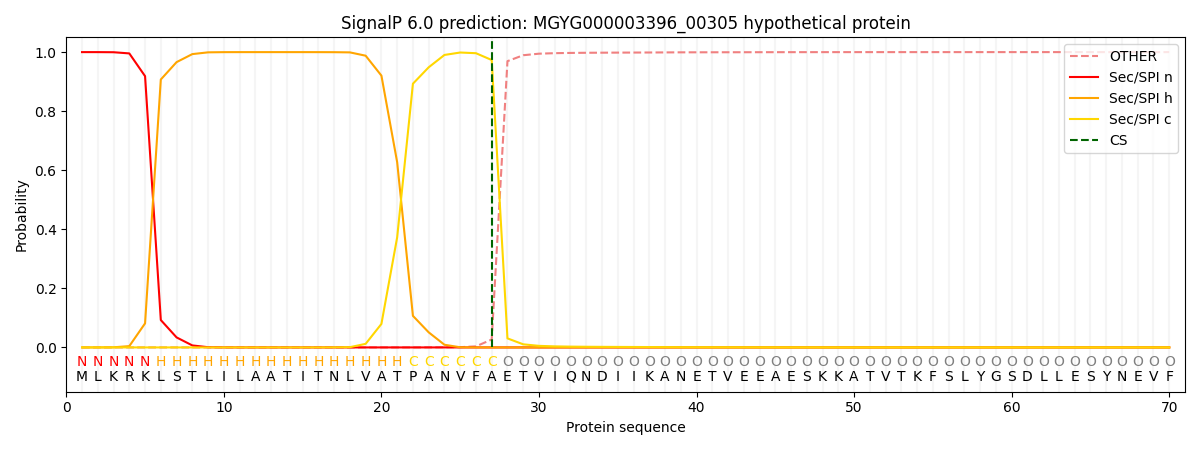
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000510 | 0.998566 | 0.000236 | 0.000243 | 0.000208 | 0.000187 |
