You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003400_01699
You are here: Home > Sequence: MGYG000003400_01699
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Stenotrophomonas sp003028475 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Xanthomonadales; Xanthomonadaceae; Stenotrophomonas; Stenotrophomonas sp003028475 | |||||||||||
| CAZyme ID | MGYG000003400_01699 | |||||||||||
| CAZy Family | GT83 | |||||||||||
| CAZyme Description | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2738; End: 4441 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT83 | 6 | 521 | 1.5e-93 | 0.9407407407407408 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1807 | ArnT | 2.94e-34 | 29 | 532 | 28 | 515 | 4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| PRK13279 | arnT | 4.60e-09 | 34 | 341 | 31 | 330 | lipid IV(A) 4-amino-4-deoxy-L-arabinosyltransferase. |
| pfam13231 | PMT_2 | 2.75e-07 | 65 | 228 | 2 | 160 | Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ARQ90946.1 | 0.0 | 1 | 567 | 1 | 567 |
| AWT15808.1 | 0.0 | 1 | 567 | 1 | 567 |
| AVO31519.1 | 0.0 | 1 | 567 | 1 | 567 |
| QHE22081.1 | 0.0 | 1 | 567 | 1 | 567 |
| SQG68535.1 | 0.0 | 1 | 567 | 1 | 567 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5EZM_A | 3.39e-18 | 15 | 340 | 33 | 358 | CrystalStructure of ArnT from Cupriavidus metallidurans in the apo state [Cupriavidus metallidurans CH34],5F15_A Crystal Structure of ArnT from Cupriavidus metallidurans bound to Undecaprenyl phosphate [Cupriavidus metallidurans CH34] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q48HY9 | 5.77e-28 | 34 | 343 | 31 | 328 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Pseudomonas savastanoi pv. phaseolicola (strain 1448A / Race 6) OX=264730 GN=arnT PE=3 SV=1 |
| Q4ZSZ0 | 2.14e-27 | 7 | 343 | 6 | 329 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Pseudomonas syringae pv. syringae (strain B728a) OX=205918 GN=arnT PE=3 SV=1 |
| C6DAW3 | 2.99e-27 | 34 | 381 | 33 | 376 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Pectobacterium carotovorum subsp. carotovorum (strain PC1) OX=561230 GN=arnT PE=3 SV=1 |
| Q4K884 | 3.51e-27 | 7 | 339 | 36 | 357 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase 2 OS=Pseudomonas fluorescens (strain ATCC BAA-477 / NRRL B-23932 / Pf-5) OX=220664 GN=arnT2 PE=3 SV=1 |
| Q3KCB9 | 2.24e-26 | 8 | 351 | 4 | 336 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase 2 OS=Pseudomonas fluorescens (strain Pf0-1) OX=205922 GN=arnT2 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.999921 | 0.000082 | 0.000009 | 0.000001 | 0.000000 | 0.000015 |
TMHMM Annotations download full data without filtering help
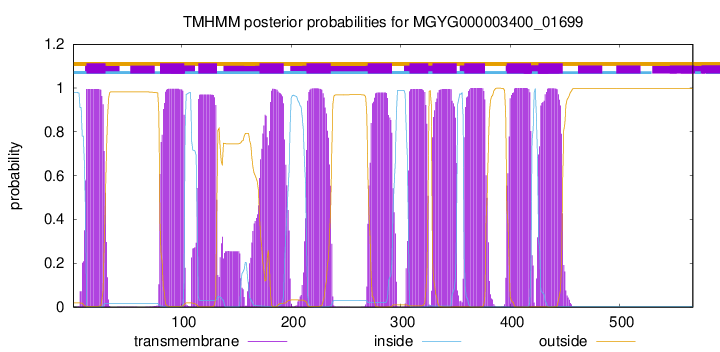
| start | end |
|---|---|
| 13 | 30 |
| 80 | 102 |
| 115 | 132 |
| 171 | 193 |
| 214 | 236 |
| 270 | 292 |
| 308 | 325 |
| 329 | 351 |
| 358 | 377 |
| 397 | 419 |
| 426 | 448 |
