You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003419_00237
You are here: Home > Sequence: MGYG000003419_00237
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | RF16 sp900766775 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Paludibacteraceae; RF16; RF16 sp900766775 | |||||||||||
| CAZyme ID | MGYG000003419_00237 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 7059; End: 8237 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PLN02773 | PLN02773 | 6.43e-11 | 19 | 341 | 23 | 247 | pectinesterase |
| PLN02990 | PLN02990 | 1.22e-10 | 182 | 340 | 363 | 502 | Probable pectinesterase/pectinesterase inhibitor |
| PLN02197 | PLN02197 | 1.93e-10 | 161 | 388 | 343 | 576 | pectinesterase |
| PLN02708 | PLN02708 | 2.65e-10 | 186 | 340 | 350 | 493 | Probable pectinesterase/pectinesterase inhibitor |
| pfam01095 | Pectinesterase | 6.63e-09 | 196 | 341 | 117 | 243 | Pectinesterase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AOW17789.1 | 7.05e-62 | 16 | 388 | 1135 | 1512 |
| QBN19519.1 | 2.47e-60 | 16 | 388 | 610 | 987 |
| AOZ99781.1 | 1.85e-59 | 16 | 388 | 1540 | 1917 |
| AUS04334.1 | 5.09e-59 | 16 | 385 | 724 | 1101 |
| AFK85374.1 | 8.54e-59 | 16 | 384 | 1335 | 1708 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5C1C_A | 3.41e-06 | 182 | 372 | 114 | 277 | CrystalStructure of the Pectin Methylesterase from Aspergillus niger in Deglycosylated Form [Aspergillus niger ATCC 1015] |
| 5C1E_A | 3.41e-06 | 182 | 372 | 114 | 277 | CrystalStructure of the Pectin Methylesterase from Aspergillus niger in Penultimately Deglycosylated Form (N-acetylglucosamine Stub at Asn84) [Aspergillus niger ATCC 1015] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8GXA1 | 4.57e-07 | 182 | 340 | 360 | 499 | Probable pectinesterase/pectinesterase inhibitor 23 OS=Arabidopsis thaliana OX=3702 GN=PME23 PE=2 SV=3 |
| O80722 | 8.17e-07 | 183 | 388 | 383 | 570 | Pectinesterase 4 OS=Arabidopsis thaliana OX=3702 GN=PME4 PE=2 SV=1 |
| Q5MFV6 | 5.84e-06 | 186 | 373 | 384 | 557 | Probable pectinesterase/pectinesterase inhibitor VGDH2 OS=Arabidopsis thaliana OX=3702 GN=VGDH2 PE=2 SV=2 |
| Q43062 | 9.67e-06 | 182 | 340 | 310 | 450 | Pectinesterase/pectinesterase inhibitor PPE8B OS=Prunus persica OX=3760 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
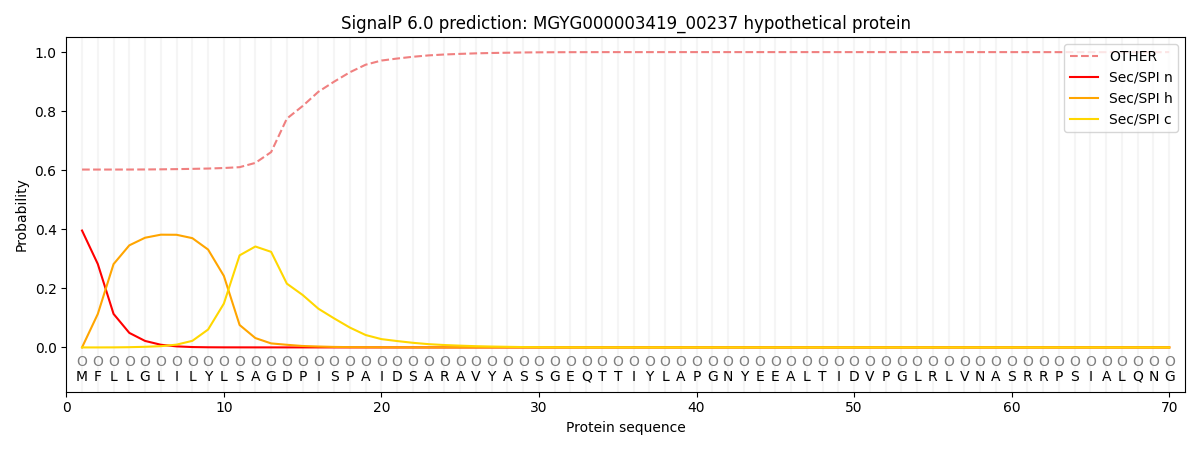
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.609155 | 0.387990 | 0.002359 | 0.000181 | 0.000144 | 0.000174 |
