You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003423_01509
You are here: Home > Sequence: MGYG000003423_01509
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
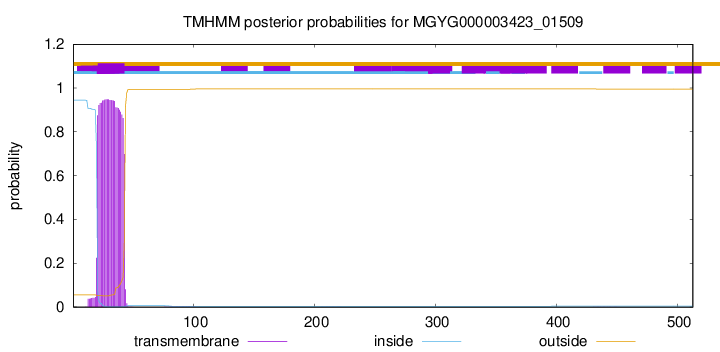
TMHMM annotations
Basic Information help
| Species | W3P20-009 sp900766825 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; W3P20-009; W3P20-009; W3P20-009 sp900766825 | |||||||||||
| CAZyme ID | MGYG000003423_01509 | |||||||||||
| CAZy Family | GH23 | |||||||||||
| CAZyme Description | Membrane-bound lytic murein transglycosylase F | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 22903; End: 24444 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH23 | 353 | 508 | 8.3e-28 | 0.8296296296296296 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd13403 | MLTF-like | 9.05e-76 | 346 | 510 | 1 | 161 | membrane-bound lytic murein transglycosylase F (MLTF) and similar proteins. This subfamily includes membrane-bound lytic murein transglycosylase F (MltF, murein lyase F) that degrades murein glycan strands. It is responsible for catalyzing the release of 1,6-anhydromuropeptides from peptidoglycan. Lytic transglycosylase catalyzes the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc) as do goose-type lysozymes. However, in addition, it also makes a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. |
| PRK10859 | PRK10859 | 1.58e-72 | 21 | 501 | 2 | 443 | membrane-bound lytic murein transglycosylase MltF. |
| COG4623 | MltF | 1.62e-60 | 61 | 501 | 15 | 424 | Membrane-bound lytic murein transglycosylase MltF [Cell wall/membrane/envelope biogenesis, Signal transduction mechanisms]. |
| cd01009 | PBP2_YfhD_N | 8.74e-34 | 69 | 319 | 1 | 221 | The solute binding domain of YfhD proteins, a member of the type 2 periplasmic binding fold protein superfamily. This subfamily includes the solute binding domain YfhD_N. These domains are found in the YfhD proteins that are predicted to function as lytic transglycosylases that cleave the glycosidic bond between N-acetylmuramic acid and N-acetylglucosamin in peptidoglycan, while the YfhD_N domain might act as an auxiliary or regulatory subunit. In addition to periplasmic solute binding domain, they have an SLT domain, typically found in soluble lytic transglycosylases, and a C-terminal low complexity domain. The YfhD proteins might have been recruited to create localized cell wall openings required for transport of large substrates such as DNA. They belong to the PBP2 superfamily of periplasmic binding proteins that differ in size and ligand specificity, but have similar tertiary structures consisting of two globular subdomains connected by a flexible hinge. They have been shown to bind their ligand in the cleft between these domains in a manner resembling a Venus flytrap. |
| pfam01464 | SLT | 2.55e-25 | 356 | 459 | 11 | 113 | Transglycosylase SLT domain. This family is distantly related to pfam00062. Members are found in phages, type II, type III and type IV secretion systems. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ASB48538.1 | 1.87e-100 | 61 | 513 | 36 | 473 |
| QZT36545.1 | 6.08e-100 | 61 | 513 | 39 | 476 |
| QZE14494.1 | 6.52e-96 | 41 | 508 | 14 | 471 |
| ALO15425.1 | 3.20e-88 | 62 | 509 | 46 | 478 |
| QCK16927.1 | 6.33e-87 | 58 | 510 | 41 | 480 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5AA2_D | 1.07e-31 | 40 | 501 | 31 | 455 | Crystalstructure of MltF from Pseudomonas aeruginosa in complex with NAM-pentapeptide. [Pseudomonas aeruginosa BWHPSA013] |
| 4OYV_A | 1.22e-31 | 62 | 501 | 11 | 421 | Crystalstructure of MltF from Pseudomonas aeruginosa complexed with leucine [Pseudomonas aeruginosa PAO1] |
| 5AA2_A | 3.65e-31 | 40 | 501 | 31 | 455 | Crystalstructure of MltF from Pseudomonas aeruginosa in complex with NAM-pentapeptide. [Pseudomonas aeruginosa BWHPSA013],5AA2_C Crystal structure of MltF from Pseudomonas aeruginosa in complex with NAM-pentapeptide. [Pseudomonas aeruginosa BWHPSA013] |
| 5AA1_A | 3.65e-31 | 40 | 501 | 31 | 455 | Crystalstructure of MltF from Pseudomonas aeruginosa in complex with NAG-anhNAM-pentapeptide [Pseudomonas aeruginosa BWHPSA013],5AA1_B Crystal structure of MltF from Pseudomonas aeruginosa in complex with NAG-anhNAM-pentapeptide [Pseudomonas aeruginosa BWHPSA013],5AA1_C Crystal structure of MltF from Pseudomonas aeruginosa in complex with NAG-anhNAM-pentapeptide [Pseudomonas aeruginosa BWHPSA013],5AA1_D Crystal structure of MltF from Pseudomonas aeruginosa in complex with NAG-anhNAM-pentapeptide [Pseudomonas aeruginosa BWHPSA013] |
| 4OZ9_A | 5.25e-31 | 62 | 501 | 4 | 414 | Crystalstructure of MltF from Pseudomonas aeruginosa complexed with isoleucine [Pseudomonas aeruginosa PAO1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A8ZWR8 | 4.93e-51 | 65 | 503 | 37 | 443 | Membrane-bound lytic murein transglycosylase F OS=Desulfococcus oleovorans (strain DSM 6200 / JCM 39069 / Hxd3) OX=96561 GN=mltF PE=3 SV=2 |
| A4SNZ5 | 4.73e-42 | 33 | 501 | 16 | 446 | Membrane-bound lytic murein transglycosylase F OS=Aeromonas salmonicida (strain A449) OX=382245 GN=mltF PE=3 SV=1 |
| Q30PQ0 | 3.61e-40 | 62 | 498 | 36 | 445 | Membrane-bound lytic murein transglycosylase F OS=Sulfurimonas denitrificans (strain ATCC 33889 / DSM 1251) OX=326298 GN=mltF PE=3 SV=2 |
| A1S850 | 8.63e-40 | 31 | 501 | 6 | 436 | Membrane-bound lytic murein transglycosylase F OS=Shewanella amazonensis (strain ATCC BAA-1098 / SB2B) OX=326297 GN=mltF PE=3 SV=2 |
| A0KJ50 | 1.50e-39 | 62 | 501 | 40 | 446 | Membrane-bound lytic murein transglycosylase F OS=Aeromonas hydrophila subsp. hydrophila (strain ATCC 7966 / DSM 30187 / BCRC 13018 / CCUG 14551 / JCM 1027 / KCTC 2358 / NCIMB 9240 / NCTC 8049) OX=380703 GN=mltF PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
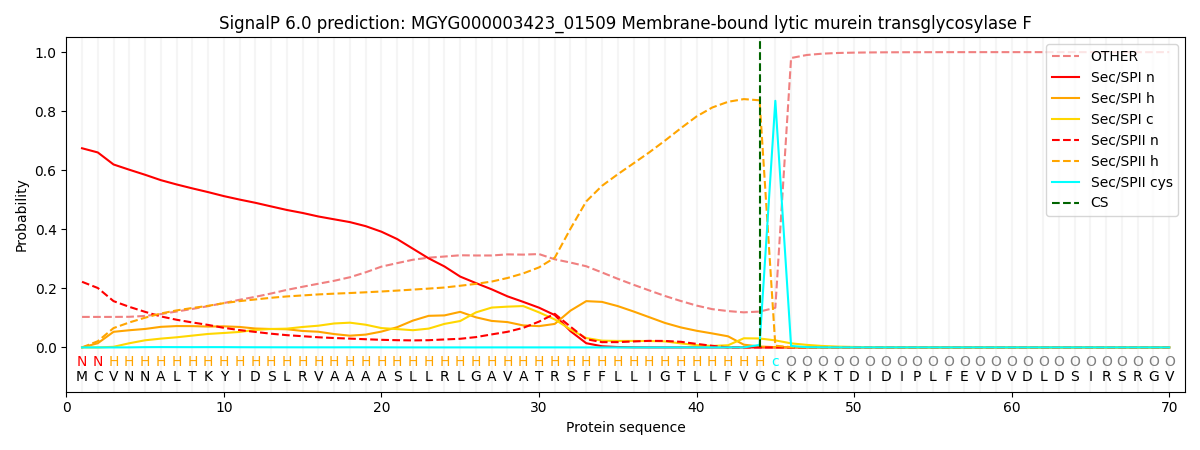
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.103134 | 0.329748 | 0.566992 | 0.000031 | 0.000076 | 0.000022 |