You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003431_00116
You are here: Home > Sequence: MGYG000003431_00116
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; HUN007; | |||||||||||
| CAZyme ID | MGYG000003431_00116 | |||||||||||
| CAZy Family | GH30 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 5632; End: 7851 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH30 | 42 | 403 | 6e-128 | 0.9912280701754386 |
| CBM22 | 474 | 604 | 5.7e-27 | 0.9694656488549618 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5520 | XynC | 6.04e-112 | 6 | 443 | 4 | 427 | O-Glycosyl hydrolase [Cell wall/membrane/envelope biogenesis]. |
| cd14256 | Dockerin_I | 7.49e-13 | 682 | 738 | 1 | 57 | Type I dockerin repeat domain. Bacterial cohesin domains bind to a complementary protein domain named dockerin, and this interaction is required for the formation of the cellulosome, a cellulose-degrading complex. The cellulosome consists of scaffoldin, a noncatalytic scaffolding polypeptide, that comprises repeating cohesion modules and a single carbohydrate-binding module (CBM). Specific calcium-dependent interactions between cohesins and dockerins appear to be essential for cellulosome assembly. This subfamily represents type I dockerins, which are responsible for anchoring a variety of enzymatic domains to the complex. |
| pfam02018 | CBM_4_9 | 2.13e-09 | 471 | 608 | 1 | 134 | Carbohydrate binding domain. This family includes diverse carbohydrate binding domains. |
| pfam00404 | Dockerin_1 | 1.06e-08 | 683 | 738 | 1 | 56 | Dockerin type I repeat. The dockerin repeat is the binding partner of the cohesin domain pfam00963. The cohesin-dockerin interaction is the crucial interaction for complex formation in the cellulosome. The dockerin repeats, each bearing homology to the EF-hand calcium-binding loop bind calcium. |
| pfam14587 | Glyco_hydr_30_2 | 3.08e-06 | 45 | 338 | 6 | 366 | O-Glycosyl hydrolase family 30. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBL17903.1 | 1.63e-230 | 3 | 627 | 2 | 599 |
| ADU21034.1 | 4.83e-226 | 39 | 633 | 7 | 579 |
| BAB39494.1 | 1.29e-225 | 39 | 633 | 36 | 608 |
| BAB39495.1 | 1.68e-165 | 173 | 633 | 1 | 451 |
| ACZ98597.1 | 5.68e-142 | 39 | 475 | 34 | 453 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3GTN_A | 3.77e-122 | 40 | 443 | 2 | 388 | CrystalStructure of XynC from Bacillus subtilis 168 [Bacillus subtilis],3GTN_B Crystal Structure of XynC from Bacillus subtilis 168 [Bacillus subtilis],3KL0_A Crystal structure of the glucuronoxylan xylanohydrolase XynC from Bacillus subtilis [Bacillus subtilis subsp. subtilis str. 168],3KL0_B Crystal structure of the glucuronoxylan xylanohydrolase XynC from Bacillus subtilis [Bacillus subtilis subsp. subtilis str. 168],3KL0_C Crystal structure of the glucuronoxylan xylanohydrolase XynC from Bacillus subtilis [Bacillus subtilis subsp. subtilis str. 168],3KL0_D Crystal structure of the glucuronoxylan xylanohydrolase XynC from Bacillus subtilis [Bacillus subtilis subsp. subtilis str. 168],3KL3_A Crystal structure of Ligand bound XynC [Bacillus subtilis subsp. subtilis str. 168],3KL3_B Crystal structure of Ligand bound XynC [Bacillus subtilis subsp. subtilis str. 168],3KL3_C Crystal structure of Ligand bound XynC [Bacillus subtilis subsp. subtilis str. 168],3KL3_D Crystal structure of Ligand bound XynC [Bacillus subtilis subsp. subtilis str. 168],3KL5_A Structure Analysis of a Xylanase From Glycosyl Hydrolase Family Thirty: Carbohydrate Ligand Complexes Reveal this Family of Enzymes Unique Mechanism of Substrate Specificity and Recognition [Bacillus subtilis],3KL5_B Structure Analysis of a Xylanase From Glycosyl Hydrolase Family Thirty: Carbohydrate Ligand Complexes Reveal this Family of Enzymes Unique Mechanism of Substrate Specificity and Recognition [Bacillus subtilis],3KL5_C Structure Analysis of a Xylanase From Glycosyl Hydrolase Family Thirty: Carbohydrate Ligand Complexes Reveal this Family of Enzymes Unique Mechanism of Substrate Specificity and Recognition [Bacillus subtilis],3KL5_D Structure Analysis of a Xylanase From Glycosyl Hydrolase Family Thirty: Carbohydrate Ligand Complexes Reveal this Family of Enzymes Unique Mechanism of Substrate Specificity and Recognition [Bacillus subtilis] |
| 4QAW_A | 1.58e-120 | 40 | 456 | 1 | 399 | Structureof modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4QAW_B Structure of modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4QAW_C Structure of modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4QAW_D Structure of modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4QAW_E Structure of modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4QAW_F Structure of modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4QAW_G Structure of modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4QAW_H Structure of modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis] |
| 4CKQ_A | 3.02e-120 | 42 | 443 | 23 | 409 | ChainA, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],4UQ9_A Chain A, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],4UQB_A Chain A, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],4UQC_A Chain A, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],4UQD_A Chain A, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],4UQE_A Chain A, Carbohydrate Binding Family 6 [Acetivibrio thermocellus] |
| 4UQA_A | 6.01e-120 | 42 | 443 | 23 | 409 | ChainA, Carbohydrate Binding Family 6 [Acetivibrio thermocellus] |
| 5A6L_A | 2.37e-119 | 42 | 443 | 23 | 409 | ChainA, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],5A6M_A Chain A, Carbohydrate Binding Family 6 [Acetivibrio thermocellus ATCC 27405] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q45070 | 1.47e-121 | 39 | 443 | 32 | 419 | Glucuronoxylanase XynC OS=Bacillus subtilis (strain 168) OX=224308 GN=xynC PE=1 SV=1 |
| Q6YK37 | 1.45e-118 | 37 | 443 | 31 | 420 | Glucuronoxylanase XynC OS=Bacillus subtilis OX=1423 GN=xynC PE=3 SV=2 |
| P51584 | 2.86e-43 | 468 | 631 | 562 | 724 | Endo-1,4-beta-xylanase Y OS=Acetivibrio thermocellus OX=1515 GN=xynY PE=1 SV=1 |
| Q53317 | 2.48e-26 | 464 | 623 | 251 | 406 | Xylanase/beta-glucanase OS=Ruminococcus flavefaciens OX=1265 GN=xynD PE=3 SV=2 |
| G2Q1N4 | 8.43e-15 | 39 | 443 | 25 | 470 | GH30 family xylanase OS=Myceliophthora thermophila (strain ATCC 42464 / BCRC 31852 / DSM 1799) OX=573729 GN=Xyn30A PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
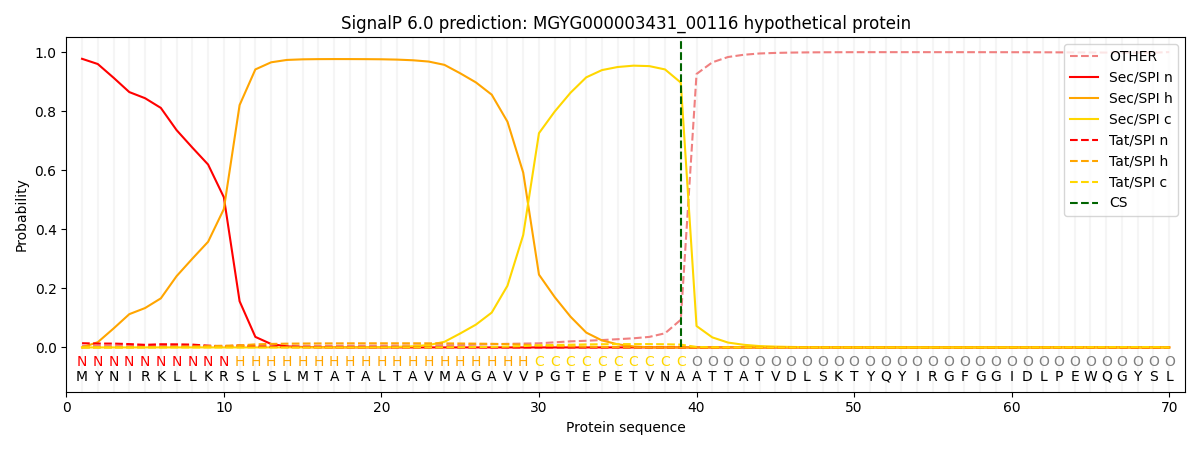
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.007301 | 0.972184 | 0.003886 | 0.015780 | 0.000518 | 0.000299 |
