You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003448_00157
You are here: Home > Sequence: MGYG000003448_00157
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | RC9 sp900767405 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; UBA932; RC9; RC9 sp900767405 | |||||||||||
| CAZyme ID | MGYG000003448_00157 | |||||||||||
| CAZy Family | GH9 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 4474; End: 6966 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH9 | 113 | 579 | 5e-74 | 0.9880382775119617 |
| CE4 | 610 | 742 | 1.7e-20 | 0.9230769230769231 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00759 | Glyco_hydro_9 | 3.20e-60 | 120 | 579 | 6 | 372 | Glycosyl hydrolase family 9. |
| cd10917 | CE4_NodB_like_6s_7s | 1.17e-32 | 614 | 812 | 1 | 171 | Catalytic NodB homology domain of rhizobial NodB-like proteins. This family belongs to the large and functionally diverse carbohydrate esterase 4 (CE4) superfamily, whose members show strong sequence similarity with some variability due to their distinct carbohydrate substrates. It includes many rhizobial NodB chitooligosaccharide N-deacetylase (EC 3.5.1.-)-like proteins, mainly from bacteria and eukaryotes, such as chitin deacetylases (EC 3.5.1.41), bacterial peptidoglycan N-acetylglucosamine deacetylases (EC 3.5.1.-), and acetylxylan esterases (EC 3.1.1.72), which catalyze the N- or O-deacetylation of substrates such as acetylated chitin, peptidoglycan, and acetylated xylan. All members of this family contain a catalytic NodB homology domain with the same overall topology and a deformed (beta/alpha)8 barrel fold with 6- or 7 strands. Their catalytic activity is dependent on the presence of a divalent cation, preferably cobalt or zinc, and they employ a conserved His-His-Asp zinc-binding triad closely associated with the conserved catalytic base (aspartic acid) and acid (histidine) to carry out acid/base catalysis. Several family members show diversity both in metal ion specificities and in the residues that coordinate the metal. |
| COG0726 | CDA1 | 4.04e-29 | 577 | 819 | 28 | 251 | Peptidoglycan/xylan/chitin deacetylase, PgdA/CDA1 family [Carbohydrate transport and metabolism, Cell wall/membrane/envelope biogenesis]. |
| cd10950 | CE4_BsYlxY_like | 3.94e-27 | 610 | 825 | 2 | 188 | Putative catalytic NodB homology domain of uncharacterized protein YlxY from Bacillus subtilis and its bacterial homologs. The Bacillus subtilis genome contains six polysaccharide deacetylase gene homologs: pdaA, pdaB (previously known as ybaN), yheN, yjeA, yxkH and ylxY. This family is represented by Bacillus subtilis putative polysaccharide deacetylase BsYlxY, encoded by the ylxY gene, which is a member of the carbohydrate esterase 4 (CE4) superfamily. Although its biological function still remains unknown, BsYlxY shows high sequence homology to the catalytic domain of Bacillus subtilis pdaB gene encoding a putative polysaccharide deacetylase (BsPdaB), which is essential for the maintenance of spores after the late stage of sporulation and is highly conserved in spore-forming bacteria. However, disruption of the ylxY gene in B. subtilis did not cause any sporulation defect. Moreover, the Asp residue in the classical His-His-Asp zinc-binding motif of CE4 esterases is mutated to a Val residue in this family. Other catalytically relevant residues of CE4 esterases are also not conserved, which suggest that members of this family may be inactive. |
| cd10949 | CE4_BsPdaB_like | 5.28e-26 | 612 | 825 | 2 | 190 | Putative catalytic NodB homology domain of Bacillus subtilis putative polysaccharide deacetylase PdaB, and its bacterial homologs. The Bacillus subtilis genome contains six polysaccharide deacetylase gene homologs: pdaA, pdaB (previously known as ybaN), yheN, yjeA, yxkH and ylxY. This family is represented by the putative polysaccharide deacetylase PdaB encoded by the pdaB gene on sporulation of Bacillus subtilis. Although its biochemical properties remain to be determined, the PdaB (YbaN) protein is essential for maintaining spores after the late stage of sporulation and is highly conserved in spore-forming bacteria. The glycans of the spore cortex may be candidate PdaB substrates. Based on sequence similarity, the family members are classified as carbohydrate esterase 4 (CE4) superfamily members. However, the classical His-His-Asp zinc-binding motif of CE4 esterases is missing in this family. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT35459.1 | 0.0 | 22 | 827 | 23 | 836 |
| QUT98069.1 | 0.0 | 22 | 827 | 23 | 836 |
| QPH59586.1 | 0.0 | 22 | 827 | 23 | 836 |
| QBJ19309.1 | 0.0 | 22 | 827 | 23 | 836 |
| QMI80662.1 | 0.0 | 22 | 827 | 23 | 836 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3X17_A | 1.30e-37 | 20 | 579 | 14 | 551 | Crystalstructure of metagenome-derived glycoside hydrolase family 9 endoglucanase [uncultured bacterium],3X17_B Crystal structure of metagenome-derived glycoside hydrolase family 9 endoglucanase [uncultured bacterium] |
| 5U2O_A | 8.29e-30 | 32 | 587 | 2 | 542 | Crystalstructure of Zn-binding triple mutant of GH family 9 endoglucanase J30 [Thermobacillus composti KWC4] |
| 6DHT_A | 4.37e-27 | 24 | 577 | 18 | 557 | Bacteroidesovatus GH9 Bacova_02649 [Bacteroides ovatus ATCC 8483] |
| 5U0H_A | 6.14e-27 | 32 | 587 | 2 | 542 | Crystalstructure of GH family 9 endoglucanase J30 [Thermobacillus composti KWC4] |
| 4M1B_A | 6.89e-18 | 606 | 825 | 49 | 243 | StructuralDetermination of BA0150, a Polysaccharide Deacetylase from Bacillus anthracis [Bacillus anthracis],4M1B_B Structural Determination of BA0150, a Polysaccharide Deacetylase from Bacillus anthracis [Bacillus anthracis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A7LXT3 | 2.69e-26 | 24 | 577 | 32 | 571 | Xyloglucan-specific endo-beta-1,4-glucanase BoGH9A OS=Bacteroides ovatus (strain ATCC 8483 / DSM 1896 / JCM 5824 / BCRC 10623 / CCUG 4943 / NCTC 11153) OX=411476 GN=BACOVA_02649 PE=1 SV=1 |
| P23658 | 4.23e-20 | 24 | 582 | 4 | 543 | Cellodextrinase OS=Butyrivibrio fibrisolvens OX=831 GN=ced1 PE=1 SV=1 |
| P14090 | 2.25e-16 | 24 | 494 | 341 | 819 | Endoglucanase C OS=Cellulomonas fimi (strain ATCC 484 / DSM 20113 / JCM 1341 / NBRC 15513 / NCIMB 8980 / NCTC 7547) OX=590998 GN=cenC PE=1 SV=2 |
| Q05156 | 2.71e-15 | 24 | 577 | 185 | 735 | Cellulase 1 OS=Streptomyces reticuli OX=1926 GN=cel1 PE=1 SV=1 |
| P50865 | 1.58e-14 | 606 | 825 | 49 | 243 | Probable polysaccharide deacetylase PdaB OS=Bacillus subtilis (strain 168) OX=224308 GN=pdaB PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
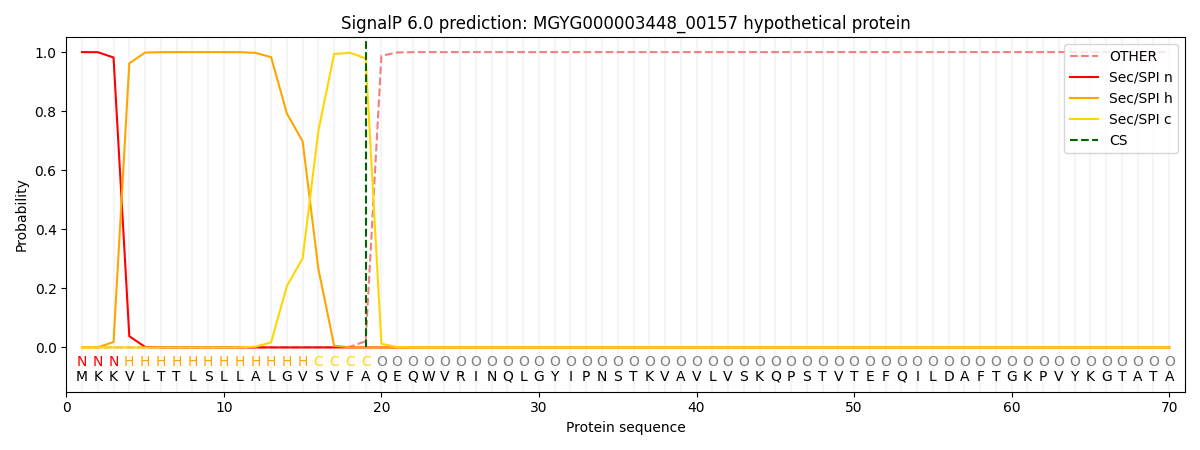
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000253 | 0.999073 | 0.000195 | 0.000158 | 0.000159 | 0.000148 |
